Junlin Yang
OpenCUA: Open Foundations for Computer-Use Agents
Aug 12, 2025Abstract:Vision-language models have demonstrated impressive capabilities as computer-use agents (CUAs) capable of automating diverse computer tasks. As their commercial potential grows, critical details of the most capable CUA systems remain closed. As these agents will increasingly mediate digital interactions and execute consequential decisions on our behalf, the research community needs access to open CUA frameworks to study their capabilities, limitations, and risks. To bridge this gap, we propose OpenCUA, a comprehensive open-source framework for scaling CUA data and foundation models. Our framework consists of: (1) an annotation infrastructure that seamlessly captures human computer-use demonstrations; (2) AgentNet, the first large-scale computer-use task dataset spanning 3 operating systems and 200+ applications and websites; (3) a scalable pipeline that transforms demonstrations into state-action pairs with reflective long Chain-of-Thought reasoning that sustain robust performance gains as data scales. Our end-to-end agent models demonstrate strong performance across CUA benchmarks. In particular, OpenCUA-32B achieves an average success rate of 34.8% on OSWorld-Verified, establishing a new state-of-the-art (SOTA) among open-source models and surpassing OpenAI CUA (GPT-4o). Further analysis confirms that our approach generalizes well across domains and benefits significantly from increased test-time computation. We release our annotation tool, datasets, code, and models to build open foundations for further CUA research.
Scaling Computer-Use Grounding via User Interface Decomposition and Synthesis
May 19, 2025Abstract:Graphical user interface (GUI) grounding, the ability to map natural language instructions to specific actions on graphical user interfaces, remains a critical bottleneck in computer use agent development. Current benchmarks oversimplify grounding tasks as short referring expressions, failing to capture the complexity of real-world interactions that require software commonsense, layout understanding, and fine-grained manipulation capabilities. To address these limitations, we introduce OSWorld-G, a comprehensive benchmark comprising 564 finely annotated samples across diverse task types including text matching, element recognition, layout understanding, and precise manipulation. Additionally, we synthesize and release the largest computer use grounding dataset Jedi, which contains 4 million examples through multi-perspective decoupling of tasks. Our multi-scale models trained on Jedi demonstrate its effectiveness by outperforming existing approaches on ScreenSpot-v2, ScreenSpot-Pro, and our OSWorld-G. Furthermore, we demonstrate that improved grounding with Jedi directly enhances agentic capabilities of general foundation models on complex computer tasks, improving from 5% to 27% on OSWorld. Through detailed ablation studies, we identify key factors contributing to grounding performance and verify that combining specialized data for different interface elements enables compositional generalization to novel interfaces. All benchmark, data, checkpoints, and code are open-sourced and available at https://osworld-grounding.github.io.
Semantic Segmentation with Generative Models: Semi-Supervised Learning and Strong Out-of-Domain Generalization
Apr 12, 2021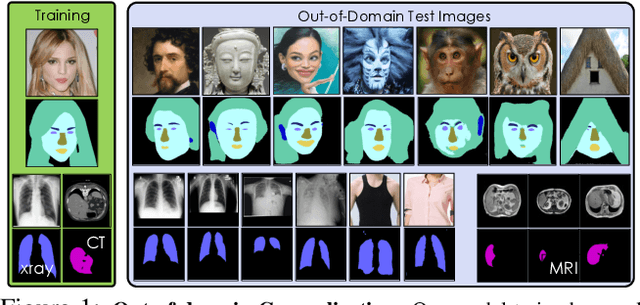
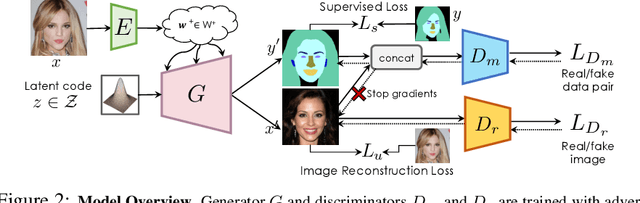
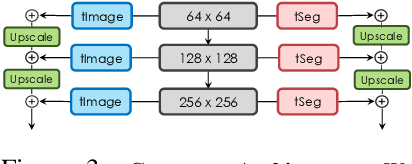
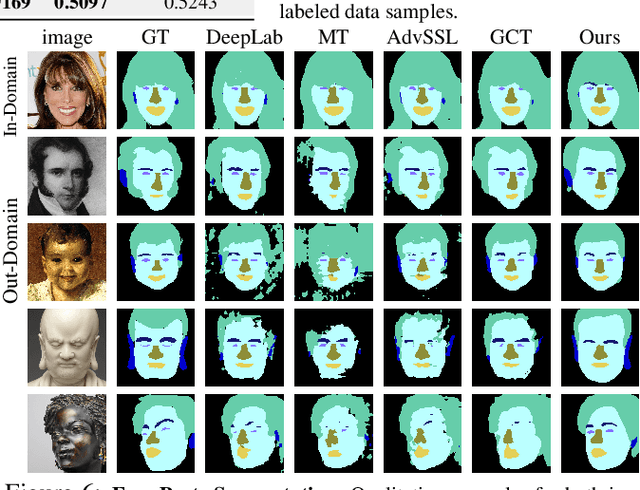
Abstract:Training deep networks with limited labeled data while achieving a strong generalization ability is key in the quest to reduce human annotation efforts. This is the goal of semi-supervised learning, which exploits more widely available unlabeled data to complement small labeled data sets. In this paper, we propose a novel framework for discriminative pixel-level tasks using a generative model of both images and labels. Concretely, we learn a generative adversarial network that captures the joint image-label distribution and is trained efficiently using a large set of unlabeled images supplemented with only few labeled ones. We build our architecture on top of StyleGAN2, augmented with a label synthesis branch. Image labeling at test time is achieved by first embedding the target image into the joint latent space via an encoder network and test-time optimization, and then generating the label from the inferred embedding. We evaluate our approach in two important domains: medical image segmentation and part-based face segmentation. We demonstrate strong in-domain performance compared to several baselines, and are the first to showcase extreme out-of-domain generalization, such as transferring from CT to MRI in medical imaging, and photographs of real faces to paintings, sculptures, and even cartoons and animal faces. Project Page: \url{https://nv-tlabs.github.io/semanticGAN/}
Unsupervised Wasserstein Distance Guided Domain Adaptation for 3D Multi-Domain Liver Segmentation
Sep 06, 2020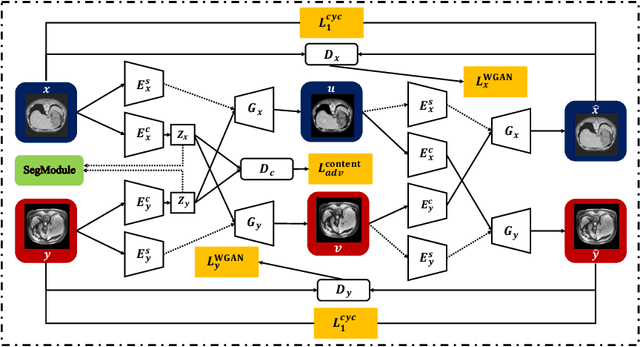
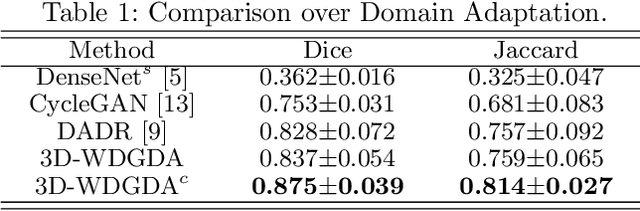
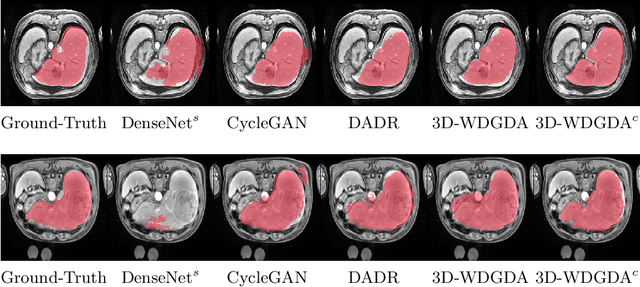
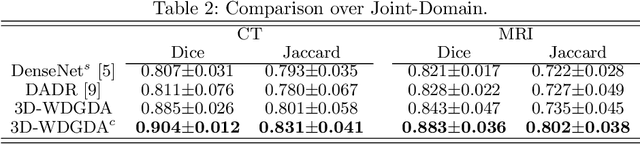
Abstract:Deep neural networks have shown exceptional learning capability and generalizability in the source domain when massive labeled data is provided. However, the well-trained models often fail in the target domain due to the domain shift. Unsupervised domain adaptation aims to improve network performance when applying robust models trained on medical images from source domains to a new target domain. In this work, we present an approach based on the Wasserstein distance guided disentangled representation to achieve 3D multi-domain liver segmentation. Concretely, we embed images onto a shared content space capturing shared feature-level information across domains and domain-specific appearance spaces. The existing mutual information-based representation learning approaches often fail to capture complete representations in multi-domain medical imaging tasks. To mitigate these issues, we utilize Wasserstein distance to learn more complete representation, and introduces a content discriminator to further facilitate the representation disentanglement. Experiments demonstrate that our method outperforms the state-of-the-art on the multi-modality liver segmentation task.
Robust and Generalizable Visual Representation Learning via Random Convolutions
Jul 25, 2020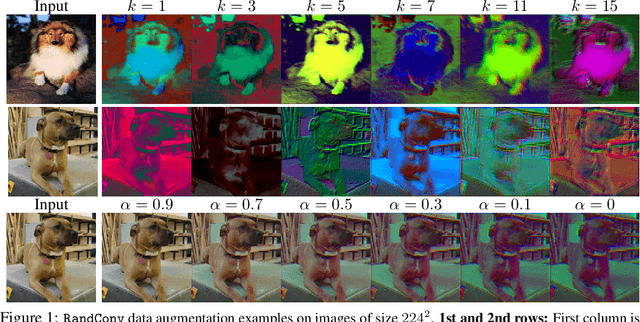
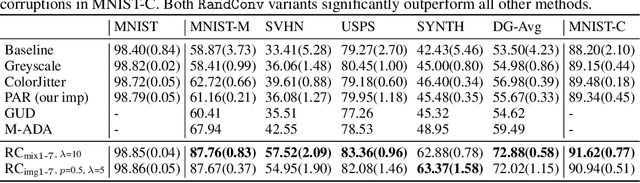
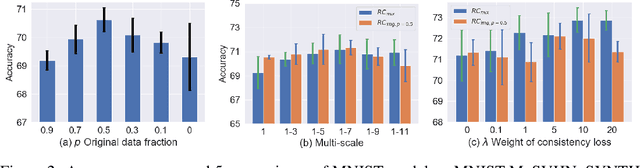
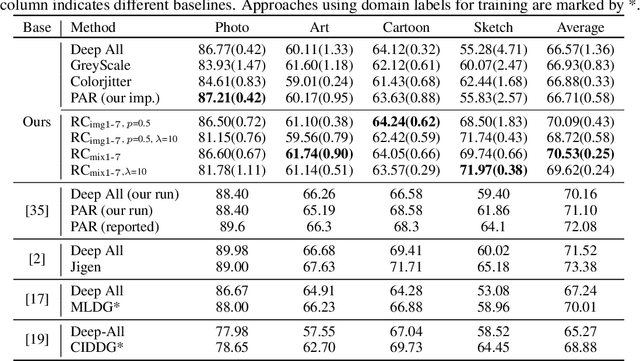
Abstract:While successful for various computer vision tasks, deep neural networks have shown to be vulnerable to texture style shifts and small perturbations to which humans are robust. Hence, our goal is to train models in such a way that improves their robustness to these perturbations. We are motivated by the approximately shape-preserving property of randomized convolutions, which is due to distance preservation under random linear transforms. Intuitively, randomized convolutions create an infinite number of new domains with similar object shapes but random local texture. Therefore, we explore using outputs of multi-scale random convolutions as new images or mixing them with the original images during training. When applying a network trained with our approach to unseen domains, our method consistently improves the performance on domain generalization benchmarks and is scalable to ImageNet. Especially for the challenging scenario of generalizing to the sketch domain in PACS and to ImageNet-Sketch, our method outperforms state-of-art methods by a large margin. More interestingly, our method can benefit downstream tasks by providing a more robust pretrained visual representation.
2018 Robotic Scene Segmentation Challenge
Feb 03, 2020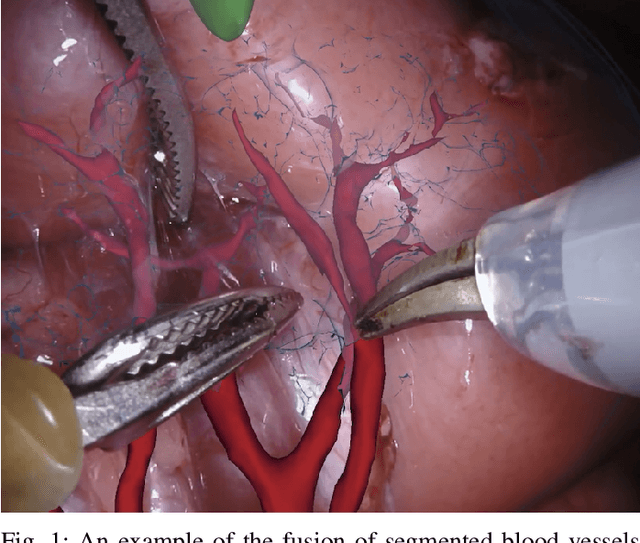
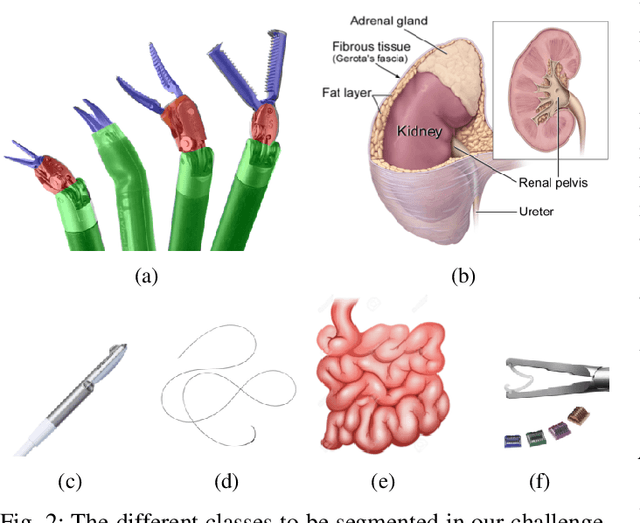
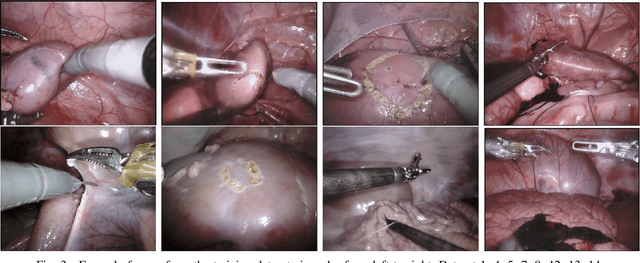
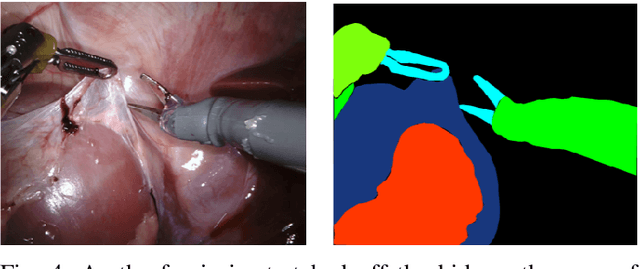
Abstract:In 2015 we began a sub-challenge at the EndoVis workshop at MICCAI in Munich using endoscope images of ex-vivo tissue with automatically generated annotations from robot forward kinematics and instrument CAD models. However, the limited background variation and simple motion rendered the dataset uninformative in learning about which techniques would be suitable for segmentation in real surgery. In 2017, at the same workshop in Quebec we introduced the robotic instrument segmentation dataset with 10 teams participating in the challenge to perform binary, articulating parts and type segmentation of da Vinci instruments. This challenge included realistic instrument motion and more complex porcine tissue as background and was widely addressed with modifications on U-Nets and other popular CNN architectures. In 2018 we added to the complexity by introducing a set of anatomical objects and medical devices to the segmented classes. To avoid over-complicating the challenge, we continued with porcine data which is dramatically simpler than human tissue due to the lack of fatty tissue occluding many organs.
Hepatocellular Carcinoma Intra-arterial Treatment Response Prediction for Improved Therapeutic Decision-Making
Dec 01, 2019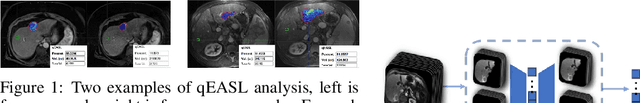
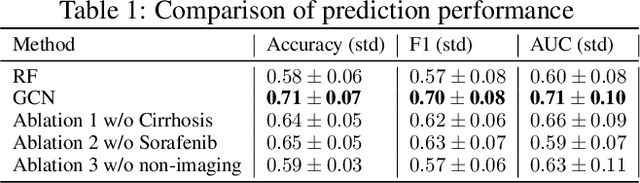
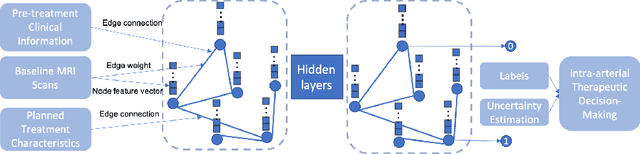
Abstract:This work proposes a pipeline to predict treatment response to intra-arterial therapy of patients with Hepatocellular Carcinoma (HCC) for improved therapeutic decision-making. Our graph neural network model seamlessly combines heterogeneous inputs of baseline MR scans, pre-treatment clinical information, and planned treatment characteristics and has been validated on patients with HCC treated by transarterial chemoembolization (TACE). It achieves Accuracy of $0.713 \pm 0.075$, F1 of $0.702 \pm 0.082$ and AUC of $0.710 \pm 0.108$. In addition, the pipeline incorporates uncertainty estimation to select hard cases and most align with the misclassified cases. The proposed pipeline arrives at more informed intra-arterial therapeutic decisions for patients with HCC via improving model accuracy and incorporating uncertainty estimation.
Decision Explanation and Feature Importance for Invertible Networks
Oct 15, 2019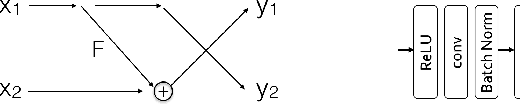

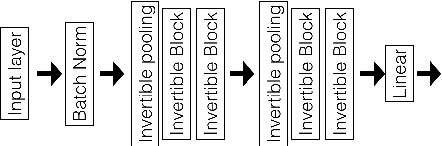

Abstract:Deep neural networks are vulnerable to adversarial attacks and hard to interpret because of their black-box nature. The recently proposed invertible network is able to accurately reconstruct the inputs to a layer from its outputs, thus has the potential to unravel the black-box model. An invertible network classifier can be viewed as a two-stage model: (1) invertible transformation from input space to the feature space; (2) a linear classifier in the feature space. We can determine the decision boundary of a linear classifier in the feature space; since the transform is invertible, we can invert the decision boundary from the feature space to the input space. Furthermore, we propose to determine the projection of a data point onto the decision boundary, and define explanation as the difference between data and its projection. Finally, we propose to locally approximate a neural network with its first-order Taylor expansion, and define feature importance using a local linear model. We provide the implementation of our method: \url{https://github.com/juntang-zhuang/explain_invertible}.
* Correct notations
Unsupervised Domain Adaptation via Disentangled Representations: Application to Cross-Modality Liver Segmentation
Aug 29, 2019
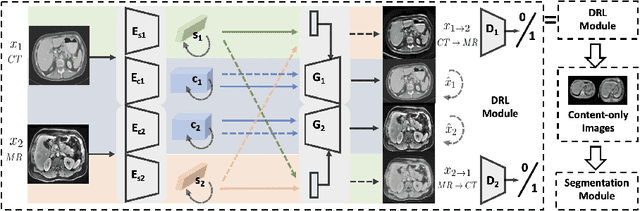
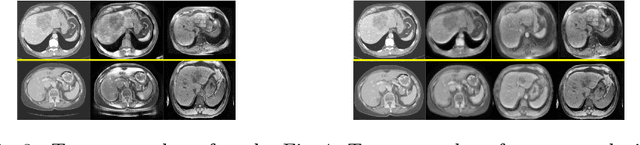
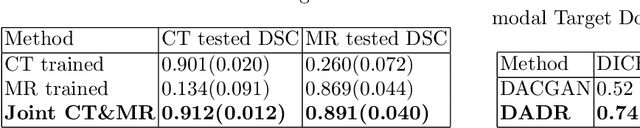
Abstract:A deep learning model trained on some labeled data from a certain source domain generally performs poorly on data from different target domains due to domain shifts. Unsupervised domain adaptation methods address this problem by alleviating the domain shift between the labeled source data and the unlabeled target data. In this work, we achieve cross-modality domain adaptation, i.e. between CT and MRI images, via disentangled representations. Compared to learning a one-to-one mapping as the state-of-art CycleGAN, our model recovers a many-to-many mapping between domains to capture the complex cross-domain relations. It preserves semantic feature-level information by finding a shared content space instead of a direct pixelwise style transfer. Domain adaptation is achieved in two steps. First, images from each domain are embedded into two spaces, a shared domain-invariant content space and a domain-specific style space. Next, the representation in the content space is extracted to perform a task. We validated our method on a cross-modality liver segmentation task, to train a liver segmentation model on CT images that also performs well on MRI. Our method achieved Dice Similarity Coefficient (DSC) of 0.81, outperforming a CycleGAN-based method of 0.72. Moreover, our model achieved good generalization to joint-domain learning, in which unpaired data from different modalities are jointly learned to improve the segmentation performance on each individual modality. Lastly, under a multi-modal target domain with significant diversity, our approach exhibited the potential for diverse image generation and remained effective with DSC of 0.74 on multi-phasic MRI while the CycleGAN-based method performed poorly with a DSC of only 0.52.
Domain-Agnostic Learning with Anatomy-Consistent Embedding for Cross-Modality Liver Segmentation
Aug 27, 2019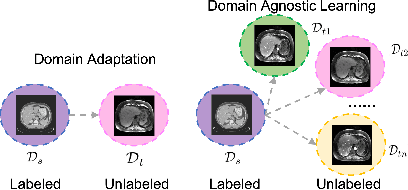
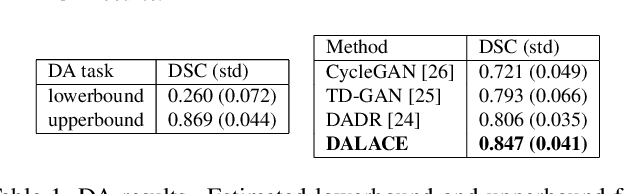
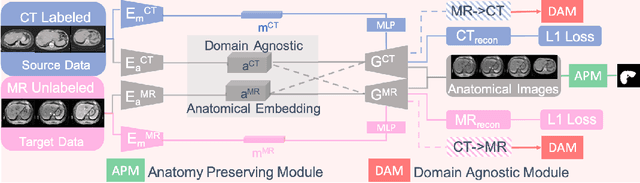
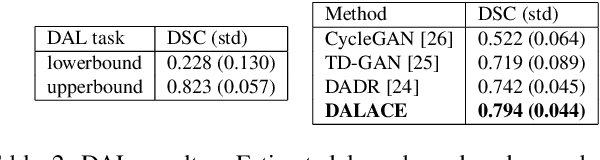
Abstract:Domain Adaptation (DA) has the potential to greatly help the generalization of deep learning models. However, the current literature usually assumes to transfer the knowledge from the source domain to a specific known target domain. Domain Agnostic Learning (DAL) proposes a new task of transferring knowledge from the source domain to data from multiple heterogeneous target domains. In this work, we propose the Domain-Agnostic Learning framework with Anatomy-Consistent Embedding (DALACE) that works on both domain-transfer and task-transfer to learn a disentangled representation, aiming to not only be invariant to different modalities but also preserve anatomical structures for the DA and DAL tasks in cross-modality liver segmentation. We validated and compared our model with state-of-the-art methods, including CycleGAN, Task Driven Generative Adversarial Network (TD-GAN), and Domain Adaptation via Disentangled Representations (DADR). For the DA task, our DALACE model outperformed CycleGAN, TD-GAN ,and DADR with DSC of 0.847 compared to 0.721, 0.793 and 0.806. For the DAL task, our model improved the performance with DSC of 0.794 from 0.522, 0.719 and 0.742 by CycleGAN, TD-GAN, and DADR. Further, we visualized the success of disentanglement, which added human interpretability of the learned meaningful representations. Through ablation analysis, we specifically showed the concrete benefits of disentanglement for downstream tasks and the role of supervision for better disentangled representation with segmentation consistency to be invariant to domains with the proposed Domain-Agnostic Module (DAM) and to preserve anatomical information with the proposed Anatomy-Preserving Module (APM).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge