Jiajie Peng
PIORS: Personalized Intelligent Outpatient Reception based on Large Language Model with Multi-Agents Medical Scenario Simulation
Nov 21, 2024



Abstract:In China, receptionist nurses face overwhelming workloads in outpatient settings, limiting their time and attention for each patient and ultimately reducing service quality. In this paper, we present the Personalized Intelligent Outpatient Reception System (PIORS). This system integrates an LLM-based reception nurse and a collaboration between LLM and hospital information system (HIS) into real outpatient reception setting, aiming to deliver personalized, high-quality, and efficient reception services. Additionally, to enhance the performance of LLMs in real-world healthcare scenarios, we propose a medical conversational data generation framework named Service Flow aware Medical Scenario Simulation (SFMSS), aiming to adapt the LLM to the real-world environments and PIORS settings. We evaluate the effectiveness of PIORS and SFMSS through automatic and human assessments involving 15 users and 15 clinical experts. The results demonstrate that PIORS-Nurse outperforms all baselines, including the current state-of-the-art model GPT-4o, and aligns with human preferences and clinical needs. Further details and demo can be found at https://github.com/FudanDISC/PIORS
CellAgent: An LLM-driven Multi-Agent Framework for Automated Single-cell Data Analysis
Jul 13, 2024Abstract:Single-cell RNA sequencing (scRNA-seq) data analysis is crucial for biological research, as it enables the precise characterization of cellular heterogeneity. However, manual manipulation of various tools to achieve desired outcomes can be labor-intensive for researchers. To address this, we introduce CellAgent (http://cell.agent4science.cn/), an LLM-driven multi-agent framework, specifically designed for the automatic processing and execution of scRNA-seq data analysis tasks, providing high-quality results with no human intervention. Firstly, to adapt general LLMs to the biological field, CellAgent constructs LLM-driven biological expert roles - planner, executor, and evaluator - each with specific responsibilities. Then, CellAgent introduces a hierarchical decision-making mechanism to coordinate these biological experts, effectively driving the planning and step-by-step execution of complex data analysis tasks. Furthermore, we propose a self-iterative optimization mechanism, enabling CellAgent to autonomously evaluate and optimize solutions, thereby guaranteeing output quality. We evaluate CellAgent on a comprehensive benchmark dataset encompassing dozens of tissues and hundreds of distinct cell types. Evaluation results consistently show that CellAgent effectively identifies the most suitable tools and hyperparameters for single-cell analysis tasks, achieving optimal performance. This automated framework dramatically reduces the workload for science data analyses, bringing us into the "Agent for Science" era.
Overview of the CAIL 2023 Argument Mining Track
Jun 20, 2024
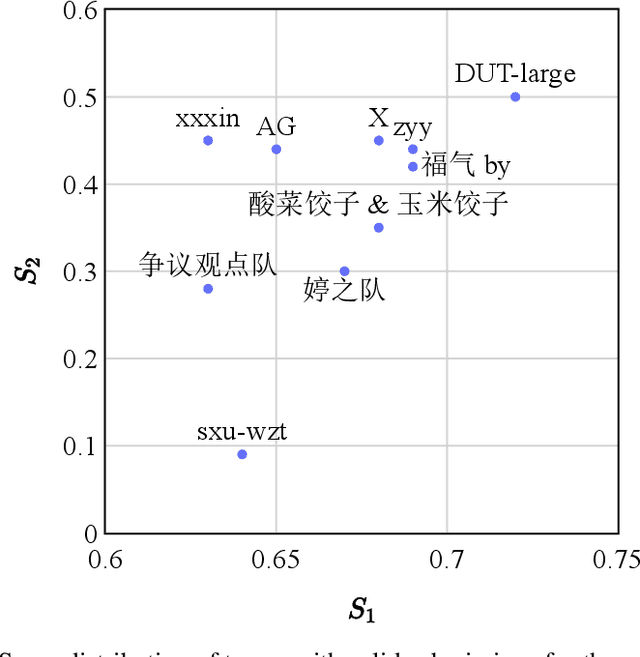
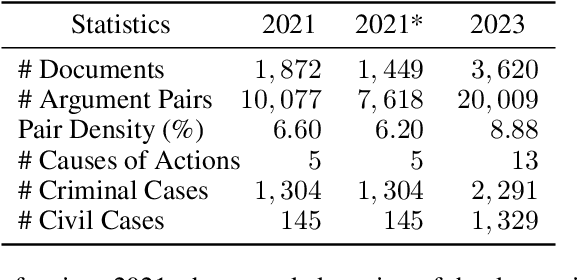
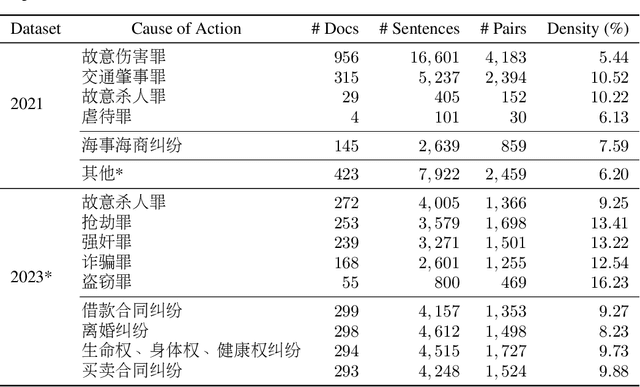
Abstract:We give a detailed overview of the CAIL 2023 Argument Mining Track, one of the Chinese AI and Law Challenge (CAIL) 2023 tracks. The main goal of the track is to identify and extract interacting argument pairs in trial dialogs. It mainly uses summarized judgment documents but can also refer to trial recordings. The track consists of two stages, and we introduce the tasks designed for each stage; we also extend the data from previous events into a new dataset -- CAIL2023-ArgMine -- with annotated new cases from various causes of action. We outline several submissions that achieve the best results, including their methods for different stages. While all submissions rely on language models, they have incorporated strategies that may benefit future work in this field.
Beyond ESM2: Graph-Enhanced Protein Sequence Modeling with Efficient Clustering
Apr 24, 2024
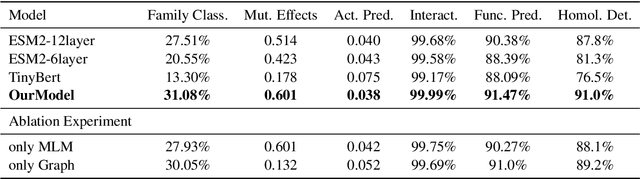


Abstract:Proteins are essential to life's processes, underpinning evolution and diversity. Advances in sequencing technology have revealed millions of proteins, underscoring the need for sophisticated pre-trained protein models for biological analysis and AI development. Facebook's ESM2, the most advanced protein language model to date, leverages a masked prediction task for unsupervised learning, crafting amino acid representations with notable biochemical accuracy. Yet, it lacks in delivering functional protein insights, signaling an opportunity for enhancing representation quality.Our study addresses this gap by incorporating protein family classification into ESM2's training.This approach, augmented with Community Propagation-Based Clustering Algorithm, improves global protein representations, while a contextual prediction task fine-tunes local amino acid accuracy. Significantly, our model achieved state-of-the-art results in several downstream experiments, demonstrating the power of combining global and local methodologies to substantially boost protein representation quality.
DISC-MedLLM: Bridging General Large Language Models and Real-World Medical Consultation
Aug 28, 2023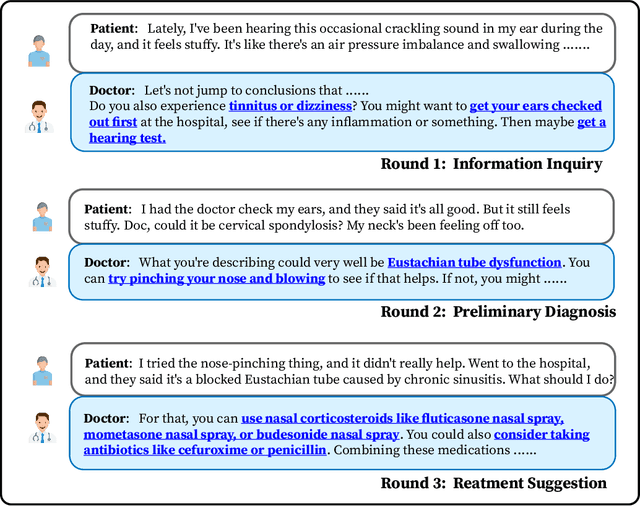
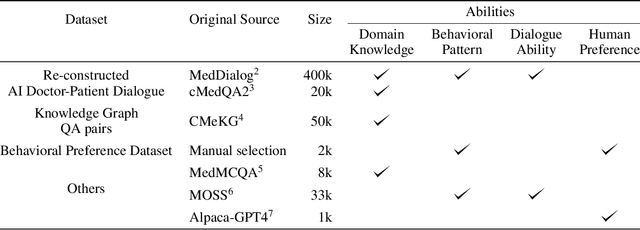
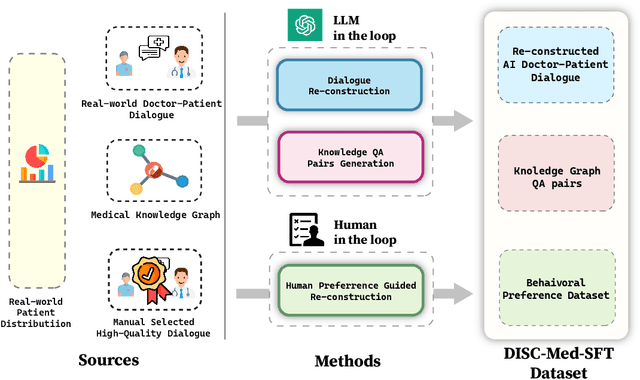
Abstract:We propose DISC-MedLLM, a comprehensive solution that leverages Large Language Models (LLMs) to provide accurate and truthful medical response in end-to-end conversational healthcare services. To construct high-quality Supervised Fine-Tuning (SFT) datasets, we employ three strategies: utilizing medical knowledge-graphs, reconstructing real-world dialogues, and incorporating human-guided preference rephrasing. These datasets are instrumental in training DISC-MedLLM, surpassing existing medical LLMs in both single-turn and multi-turn consultation scenarios. Extensive experimental results demonstrate the effectiveness of the proposed model in bridging the gap between general language models and real-world medical consultation. Additionally, we release the constructed dataset and model weights to further contribute to research and development. Further details and resources can be found at https://github.com/FudanDISC/DISC-MedLLM
DxFormer: A Decoupled Automatic Diagnostic System Based on Decoder-Encoder Transformer with Dense Symptom Representations
May 08, 2022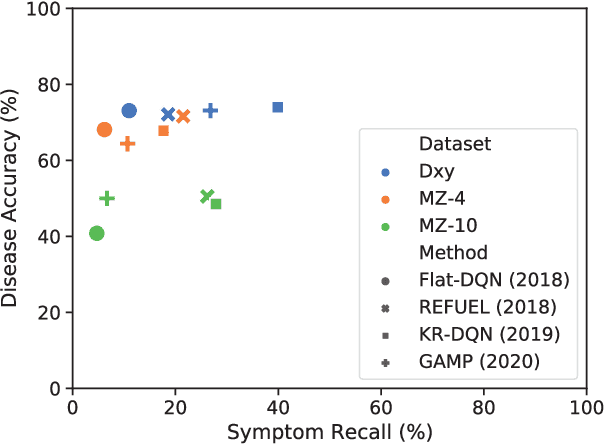
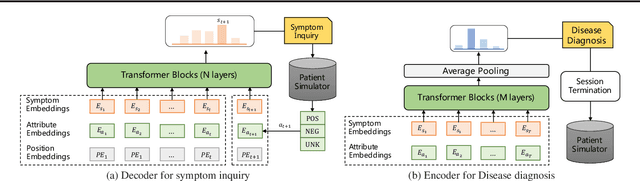
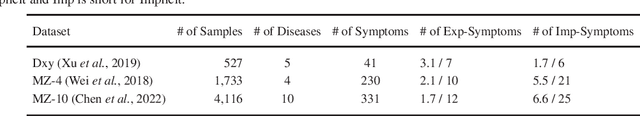
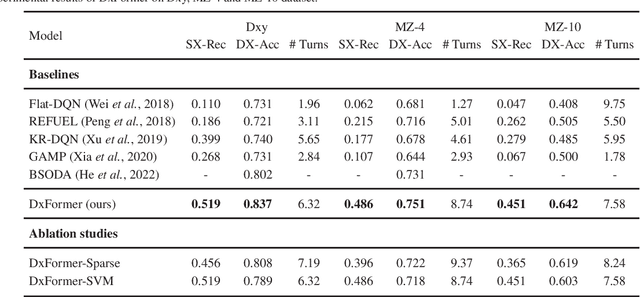
Abstract:Diagnosis-oriented dialogue system queries the patient's health condition and makes predictions about possible diseases through continuous interaction with the patient. A few studies use reinforcement learning (RL) to learn the optimal policy from the joint action space of symptoms and diseases. However, existing RL (or Non-RL) methods cannot achieve sufficiently good prediction accuracy, still far from its upper limit. To address the problem, we propose a decoupled automatic diagnostic framework DxFormer, which divides the diagnosis process into two steps: symptom inquiry and disease diagnosis, where the transition from symptom inquiry to disease diagnosis is explicitly determined by the stopping criteria. In DxFormer, we treat each symptom as a token, and formalize the symptom inquiry and disease diagnosis to a language generation model and a sequence classification model respectively. We use the inverted version of Transformer, i.e., the decoder-encoder structure, to learn the representation of symptoms by jointly optimizing the reinforce reward and cross entropy loss. Extensive experiments on three public real-world datasets prove that our proposed model can effectively learn doctors' clinical experience and achieve the state-of-the-art results in terms of symptom recall and diagnostic accuracy.
A Benchmark for Automatic Medical Consultation System: Frameworks, Tasks and Datasets
Apr 27, 2022
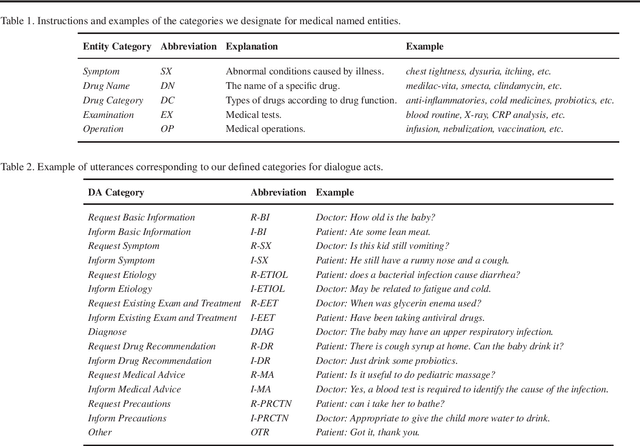
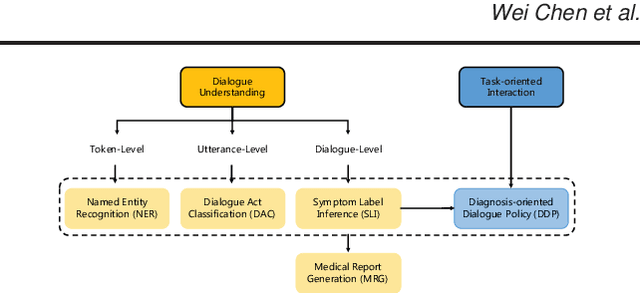
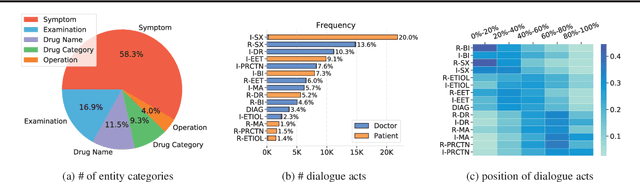
Abstract:In recent years, interest has arisen in using machine learning to improve the efficiency of automatic medical consultation and enhance patient experience. In this paper, we propose two frameworks to support automatic medical consultation, namely doctor-patient dialogue understanding and task-oriented interaction. A new large medical dialogue dataset with multi-level fine-grained annotations is introduced and five independent tasks are established, including named entity recognition, dialogue act classification, symptom label inference, medical report generation and diagnosis-oriented dialogue policy. We report a set of benchmark results for each task, which shows the usability of the dataset and sets a baseline for future studies.
ED2: An Environment Dynamics Decomposition Framework for World Model Construction
Dec 06, 2021



Abstract:Model-based reinforcement learning methods achieve significant sample efficiency in many tasks, but their performance is often limited by the existence of the model error. To reduce the model error, previous works use a single well-designed network to fit the entire environment dynamics, which treats the environment dynamics as a black box. However, these methods lack to consider the environmental decomposed property that the dynamics may contain multiple sub-dynamics, which can be modeled separately, allowing us to construct the world model more accurately. In this paper, we propose the Environment Dynamics Decomposition (ED2), a novel world model construction framework that models the environment in a decomposing manner. ED2 contains two key components: sub-dynamics discovery (SD2) and dynamics decomposition prediction (D2P). SD2 discovers the sub-dynamics in an environment and then D2P constructs the decomposed world model following the sub-dynamics. ED2 can be easily combined with existing MBRL algorithms and empirical results show that ED2 significantly reduces the model error and boosts the performance of the state-of-the-art MBRL algorithms on various tasks.
Curriculum Learning for Vision-and-Language Navigation
Nov 14, 2021



Abstract:Vision-and-Language Navigation (VLN) is a task where an agent navigates in an embodied indoor environment under human instructions. Previous works ignore the distribution of sample difficulty and we argue that this potentially degrade their agent performance. To tackle this issue, we propose a novel curriculum-based training paradigm for VLN tasks that can balance human prior knowledge and agent learning progress about training samples. We develop the principle of curriculum design and re-arrange the benchmark Room-to-Room (R2R) dataset to make it suitable for curriculum training. Experiments show that our method is model-agnostic and can significantly improve the performance, the generalizability, and the training efficiency of current state-of-the-art navigation agents without increasing model complexity.
Efficient Deep Reinforcement Learning through Policy Transfer
Feb 19, 2020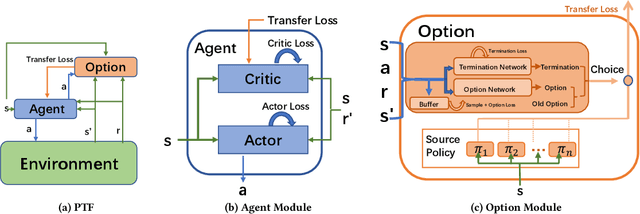
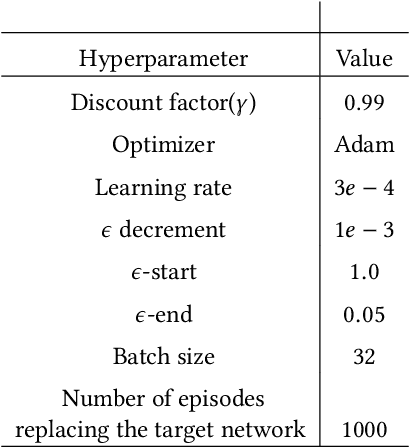
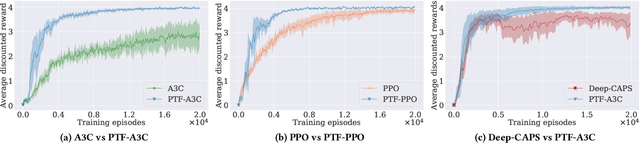
Abstract:Transfer Learning (TL) has shown great potential to accelerate Reinforcement Learning (RL) by leveraging prior knowledge from past learned policies of relevant tasks. Existing transfer approaches either explicitly computes the similarity between tasks or select appropriate source policies to provide guided explorations for the target task. However, how to directly optimize the target policy by alternatively utilizing knowledge from appropriate source policies without explicitly measuring the similarity is currently missing. In this paper, we propose a novel Policy Transfer Framework (PTF) to accelerate RL by taking advantage of this idea. Our framework learns when and which source policy is the best to reuse for the target policy and when to terminate it by modeling multi-policy transfer as the option learning problem. PTF can be easily combined with existing deep RL approaches. Experimental results show it significantly accelerates the learning process and surpasses state-of-the-art policy transfer methods in terms of learning efficiency and final performance in both discrete and continuous action spaces.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge