Guojing Cong
Oak Ridge National Laboratory
Predicting Drug Effects from High-Dimensional, Asymmetric Drug Datasets by Using Graph Neural Networks: A Comprehensive Analysis of Multitarget Drug Effect Prediction
Oct 11, 2024



Abstract:Graph neural networks (GNNs) have emerged as one of the most effective ML techniques for drug effect prediction from drug molecular graphs. Despite having immense potential, GNN models lack performance when using datasets that contain high-dimensional, asymmetrically co-occurrent drug effects as targets with complex correlations between them. Training individual learning models for each drug effect and incorporating every prediction result for a wide spectrum of drug effects are impractical. Therefore, an opportunity exists to address this challenge as multitarget prediction problems and predict all drug effects at a time. We developed standard and hybrid GNNs to perform two separate tasks: multiregression for continuous values and multilabel classification for categorical values contained in our datasets. Because multilabel classification makes the target data even more sparse and introduces asymmetric label co-occurrence, learning these models becomes difficult and heavily impacts the GNN's performance. To address these challenges, we propose a new data oversampling technique to improve multilabel classification performances on all the given imbalanced molecular graph datasets. Using the technique, we improve the data imbalance ratio of the drug effects while protecting the datasets' integrity. Finally, we evaluate the multilabel classification performance of the best-performing hybrid GNN model on all the oversampled datasets obtained from the proposed oversampling technique. In all the evaluation metrics (i.e., precision, recall, and F1 score), this model significantly outperforms other ML models, including GNN models when they are trained on the original datasets or oversampled datasets with MLSMOTE, which is a well-known oversampling technique.
Transductive Spiking Graph Neural Networks for Loihi
Apr 25, 2024



Abstract:Graph neural networks have emerged as a specialized branch of deep learning, designed to address problems where pairwise relations between objects are crucial. Recent advancements utilize graph convolutional neural networks to extract features within graph structures. Despite promising results, these methods face challenges in real-world applications due to sparse features, resulting in inefficient resource utilization. Recent studies draw inspiration from the mammalian brain and employ spiking neural networks to model and learn graph structures. However, these approaches are limited to traditional Von Neumann-based computing systems, which still face hardware inefficiencies. In this study, we present a fully neuromorphic implementation of spiking graph neural networks designed for Loihi 2. We optimize network parameters using Lava Bayesian Optimization, a novel hyperparameter optimization system compatible with neuromorphic computing architectures. We showcase the performance benefits of combining neuromorphic Bayesian optimization with our approach for citation graph classification using fixed-precision spiking neurons. Our results demonstrate the capability of integer-precision, Loihi 2 compatible spiking neural networks in performing citation graph classification with comparable accuracy to existing floating point implementations.
Optimizing Distributed Training on Frontier for Large Language Models
Dec 21, 2023
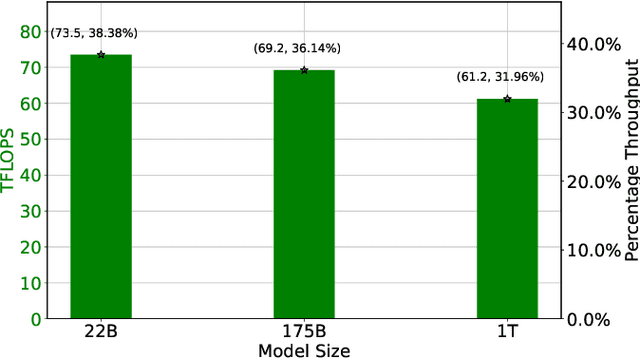
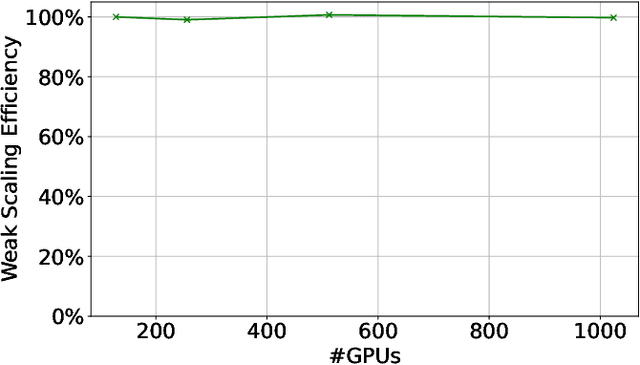
Abstract:Large language models (LLMs) have demonstrated remarkable success as foundational models, benefiting various downstream applications through fine-tuning. Recent studies on loss scaling have demonstrated the superior performance of larger LLMs compared to their smaller counterparts. Nevertheless, training LLMs with billions of parameters poses significant challenges and requires considerable computational resources. For example, training a one trillion parameter GPT-style model on 20 trillion tokens requires a staggering 120 million exaflops of computation. This research explores efficient distributed training strategies to extract this computation from Frontier, the world's first exascale supercomputer dedicated to open science. We enable and investigate various model and data parallel training techniques, such as tensor parallelism, pipeline parallelism, and sharded data parallelism, to facilitate training a trillion-parameter model on Frontier. We empirically assess these techniques and their associated parameters to determine their impact on memory footprint, communication latency, and GPU's computational efficiency. We analyze the complex interplay among these techniques and find a strategy to combine them to achieve high throughput through hyperparameter tuning. We have identified efficient strategies for training large LLMs of varying sizes through empirical analysis and hyperparameter tuning. For 22 Billion, 175 Billion, and 1 Trillion parameters, we achieved GPU throughputs of $38.38\%$, $36.14\%$, and $31.96\%$, respectively. For the training of the 175 Billion parameter model and the 1 Trillion parameter model, we achieved $100\%$ weak scaling efficiency on 1024 and 3072 MI250X GPUs, respectively. We also achieved strong scaling efficiencies of $89\%$ and $87\%$ for these two models.
DeepSpeed4Science Initiative: Enabling Large-Scale Scientific Discovery through Sophisticated AI System Technologies
Oct 11, 2023



Abstract:In the upcoming decade, deep learning may revolutionize the natural sciences, enhancing our capacity to model and predict natural occurrences. This could herald a new era of scientific exploration, bringing significant advancements across sectors from drug development to renewable energy. To answer this call, we present DeepSpeed4Science initiative (deepspeed4science.ai) which aims to build unique capabilities through AI system technology innovations to help domain experts to unlock today's biggest science mysteries. By leveraging DeepSpeed's current technology pillars (training, inference and compression) as base technology enablers, DeepSpeed4Science will create a new set of AI system technologies tailored for accelerating scientific discoveries by addressing their unique complexity beyond the common technical approaches used for accelerating generic large language models (LLMs). In this paper, we showcase the early progress we made with DeepSpeed4Science in addressing two of the critical system challenges in structural biology research.
AI-aided multiscale modeling of physiologically-significant blood clots
May 25, 2022Abstract:We have developed an AI-aided multiple time stepping (AI-MTS) algorithm and multiscale modeling framework (AI-MSM) and implemented them on the Summit-like supercomputer, AIMOS. AI-MSM is the first of its kind to integrate multi-physics, including intra-platelet, inter-platelet, and fluid-platelet interactions, into one system. It has simulated a record-setting multiscale blood clotting model of 102 million particles, of which 70 flowing and 180 aggregating platelets, under dissipative particle dynamics to coarse-grained molecular dynamics. By adaptively adjusting timestep sizes to match the characteristic time scales of the underlying dynamics, AI-MTS optimally balances speeds and accuracies of the simulations.
Accelerate Distributed Stochastic Descent for Nonconvex Optimization with Momentum
Oct 01, 2021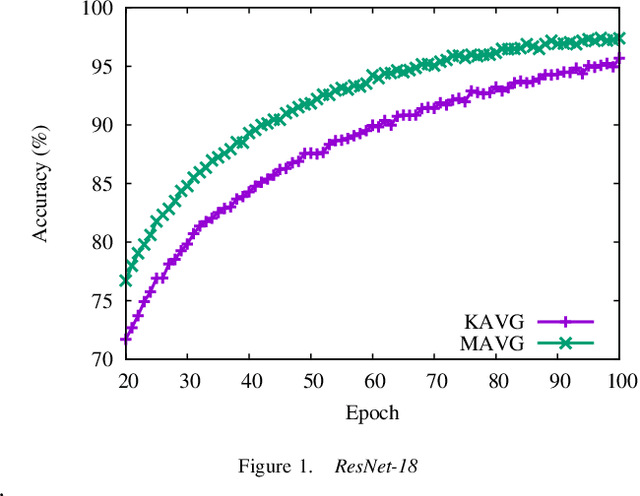
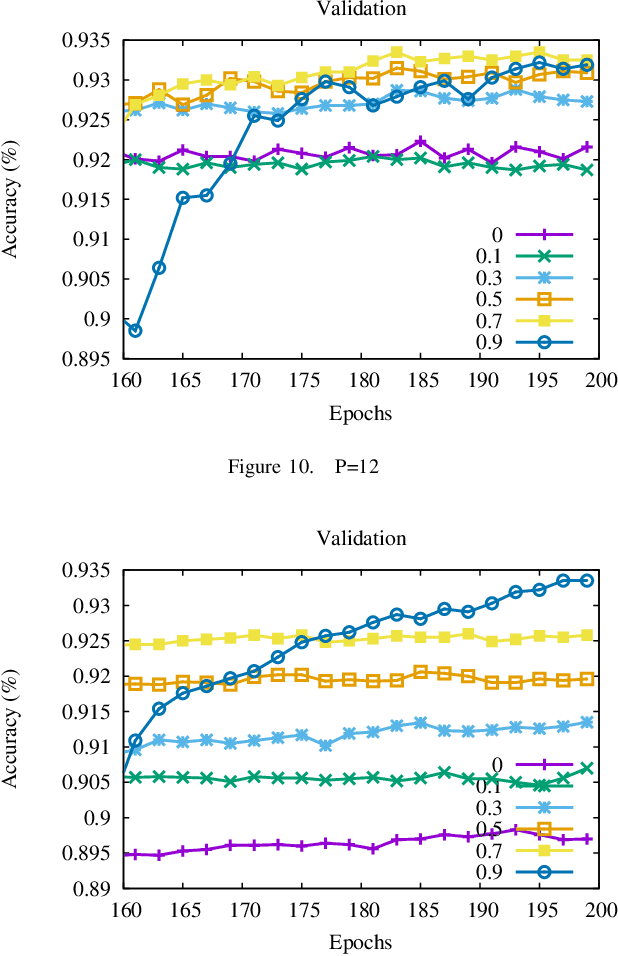
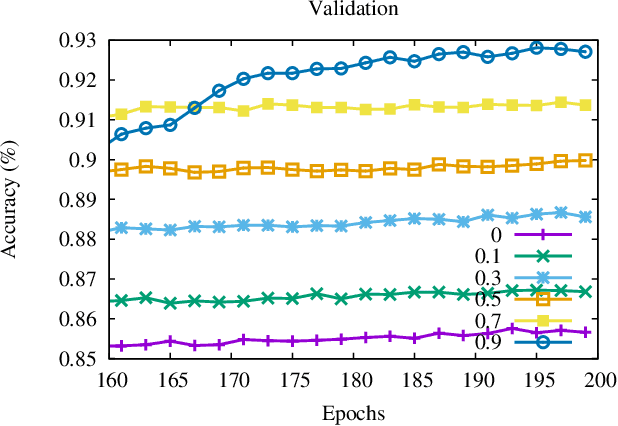
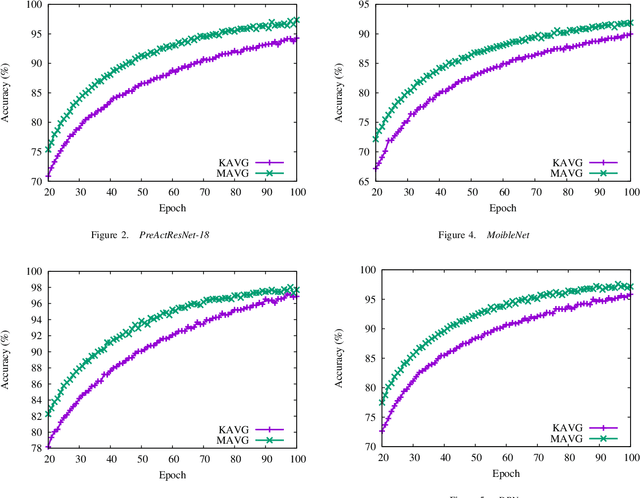
Abstract:Momentum method has been used extensively in optimizers for deep learning. Recent studies show that distributed training through K-step averaging has many nice properties. We propose a momentum method for such model averaging approaches. At each individual learner level traditional stochastic gradient is applied. At the meta-level (global learner level), one momentum term is applied and we call it block momentum. We analyze the convergence and scaling properties of such momentum methods. Our experimental results show that block momentum not only accelerates training, but also achieves better results.
CASTELO: Clustered Atom Subtypes aidEd Lead Optimization -- a combined machine learning and molecular modeling method
Nov 27, 2020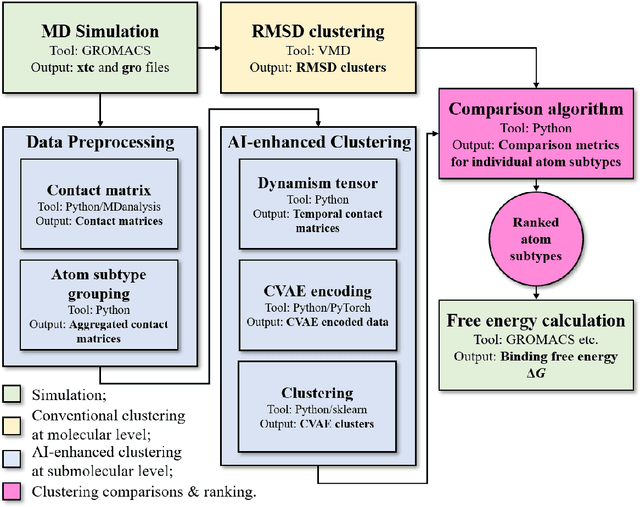

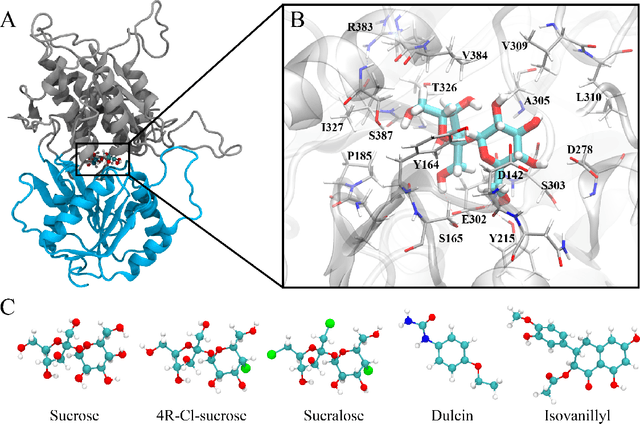
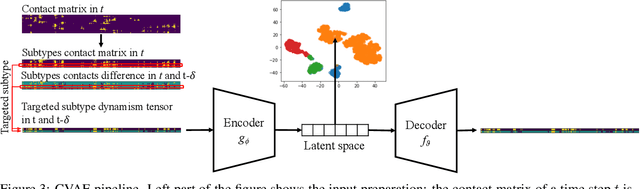
Abstract:Drug discovery is a multi-stage process that comprises two costly major steps: pre-clinical research and clinical trials. Among its stages, lead optimization easily consumes more than half of the pre-clinical budget. We propose a combined machine learning and molecular modeling approach that automates lead optimization workflow \textit{in silico}. The initial data collection is achieved with physics-based molecular dynamics (MD) simulation. Contact matrices are calculated as the preliminary features extracted from the simulations. To take advantage of the temporal information from the simulations, we enhanced contact matrices data with temporal dynamism representation, which are then modeled with unsupervised convolutional variational autoencoder (CVAE). Finally, conventional clustering method and CVAE-based clustering method are compared with metrics to rank the submolecular structures and propose potential candidates for lead optimization. With no need for extensive structure-activity relationship database, our method provides new hints for drug modification hotspots which can be used to improve drug efficacy. Our workflow can potentially reduce the lead optimization turnaround time from months/years to days compared with the conventional labor-intensive process and thus can potentially become a valuable tool for medical researchers.
Accelerating Data Loading in Deep Neural Network Training
Oct 02, 2019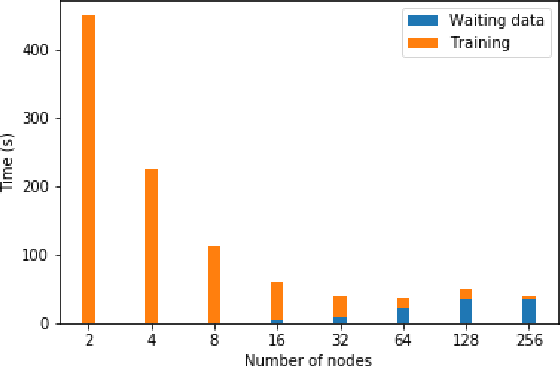
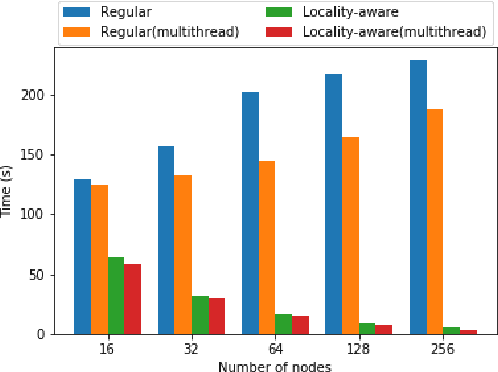
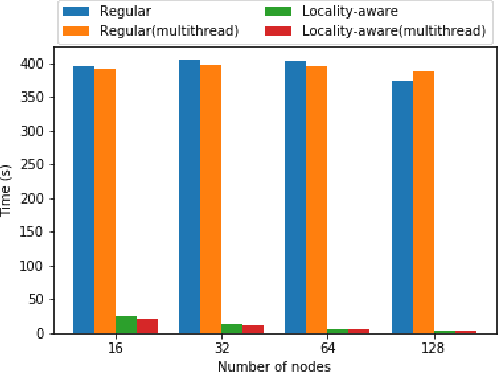
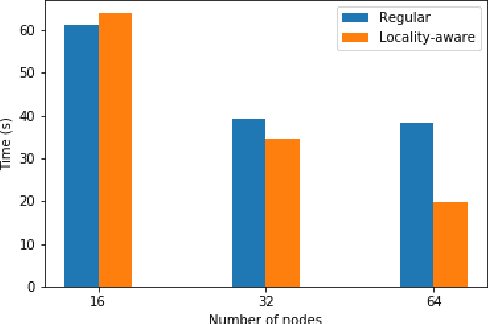
Abstract:Data loading can dominate deep neural network training time on large-scale systems. We present a comprehensive study on accelerating data loading performance in large-scale distributed training. We first identify performance and scalability issues in current data loading implementations. We then propose optimizations that utilize CPU resources to the data loader design. We use an analytical model to characterize the impact of data loading on the overall training time and establish the performance trend as we scale up distributed training. Our model suggests that I/O rate limits the scalability of distributed training, which inspires us to design a locality-aware data loading method. By utilizing software caches, our method can drastically reduce the data loading communication volume in comparison with the original data loading implementation. Finally, we evaluate the proposed optimizations with various experiments. We achieved more than 30x speedup in data loading using 256 nodes with 1,024 learners.
A Distributed Hierarchical SGD Algorithm with Sparse Global Reduction
Mar 12, 2019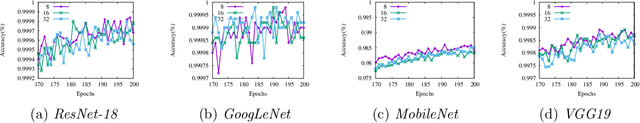
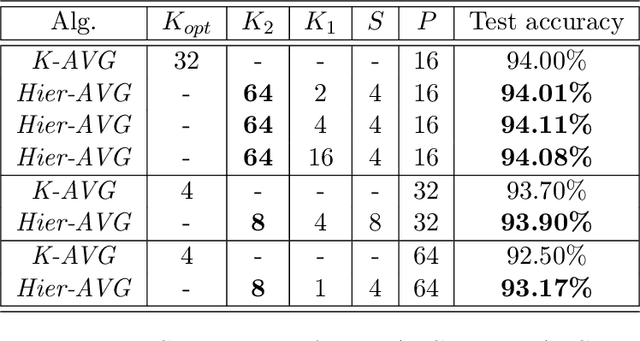
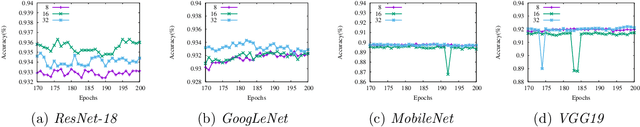
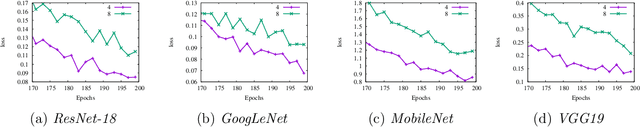
Abstract:Reducing communication overhead is a big challenge for large-scale distributed training. To address this issue, we present a hierarchical averaging stochastic gradient descent (Hier-AVG) algorithm that reduces global reductions (averaging) by employing less costly local reductions. As a very general type of parallel SGD, Hier-AVG can reproduce several commonly adopted synchronous parallel SGD variants by adjusting its parameters. We establish standard convergence results of Hier-AVG for non-convex smooth optimization problems. Under the non-asymptotic scenario, we show that Hier-AVG with less frequent global averaging can sometimes have faster training speed. In addition, we show that more frequent local averaging with more participants involved can lead to faster training convergence. By comparing Hier-AVG with another distributed training algorithm K-AVG, we show that through deploying local averaging with less global averaging Hier-AVG can still achieve comparable training speed while constantly get better test accuracy. As a result, local averaging can serve as an alternative remedy to effectively reduce communication overhead when the number of learners is large. We test Hier-AVG with several state-of-the-art deep neural nets on CIFAR-10 to validate our analysis. Further experiments to compare Hier-AVG with K-AVG on ImageNet-1K also show Hier-AVG's superiority over K-AVG.
On the convergence properties of a $K$-step averaging stochastic gradient descent algorithm for nonconvex optimization
May 16, 2018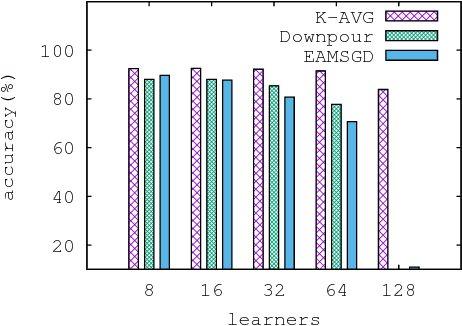
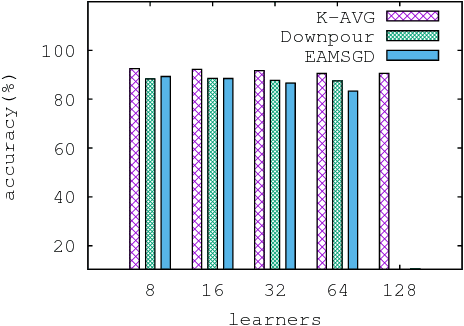
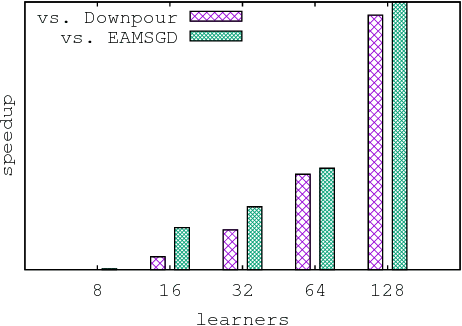
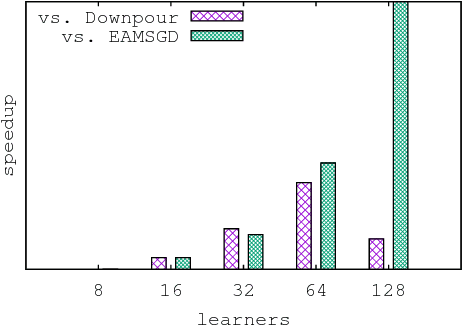
Abstract:Despite their popularity, the practical performance of asynchronous stochastic gradient descent methods (ASGD) for solving large scale machine learning problems are not as good as theoretical results indicate. We adopt and analyze a synchronous K-step averaging stochastic gradient descent algorithm which we call K-AVG. We establish the convergence results of K-AVG for nonconvex objectives and explain why the K-step delay is necessary and leads to better performance than traditional parallel stochastic gradient descent which is a special case of K-AVG with $K=1$. We also show that K-AVG scales better than ASGD. Another advantage of K-AVG over ASGD is that it allows larger stepsizes. On a cluster of $128$ GPUs, K-AVG is faster than ASGD implementations and achieves better accuracies and faster convergence for \cifar dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge