Sajal Dash
Scalable Artificial Intelligence for Science: Perspectives, Methods and Exemplars
Jun 24, 2024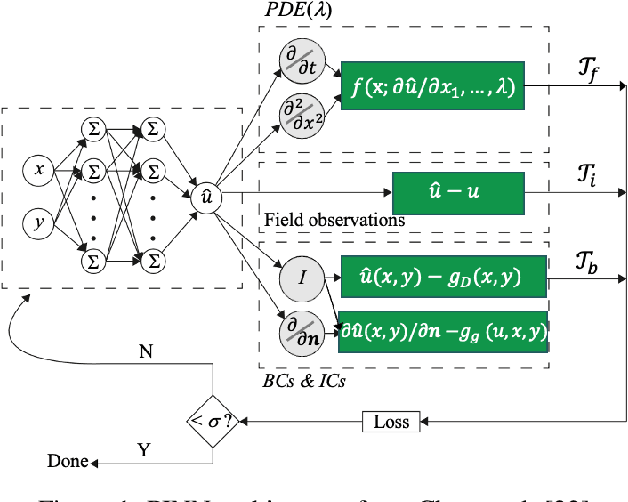
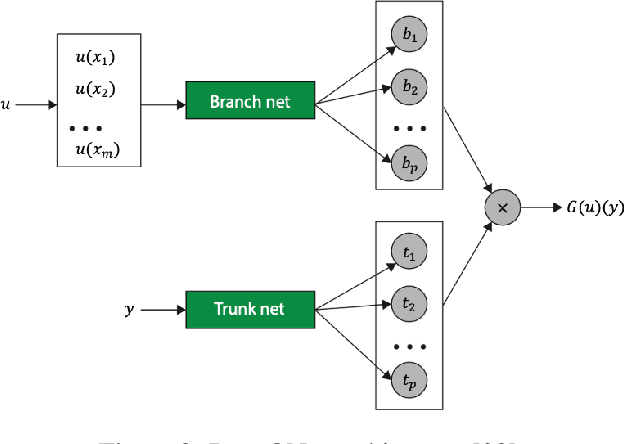
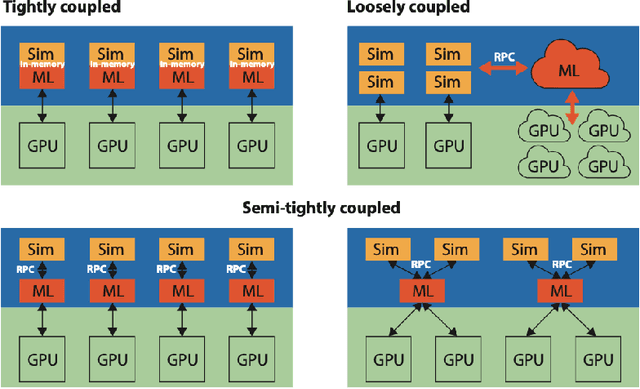
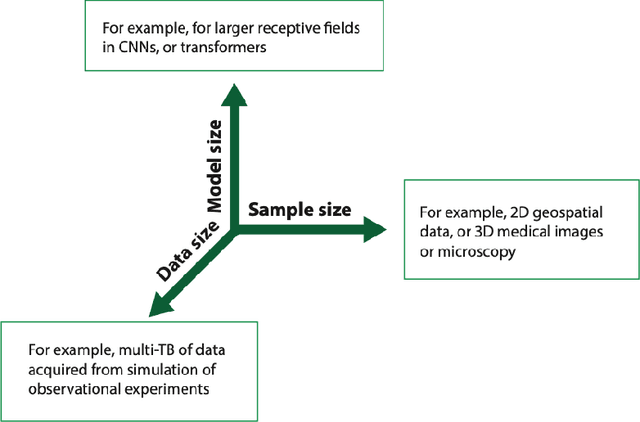
Abstract:In a post-ChatGPT world, this paper explores the potential of leveraging scalable artificial intelligence for scientific discovery. We propose that scaling up artificial intelligence on high-performance computing platforms is essential to address such complex problems. This perspective focuses on scientific use cases like cognitive simulations, large language models for scientific inquiry, medical image analysis, and physics-informed approaches. The study outlines the methodologies needed to address such challenges at scale on supercomputers or the cloud and provides exemplars of such approaches applied to solve a variety of scientific problems.
Optimizing Distributed Training on Frontier for Large Language Models
Dec 21, 2023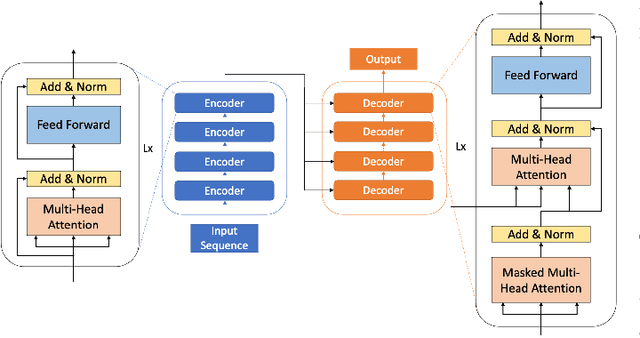
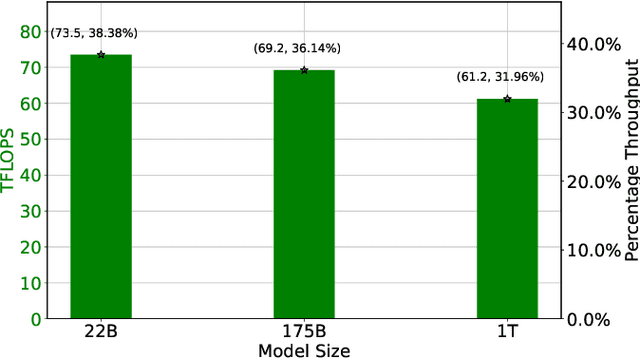
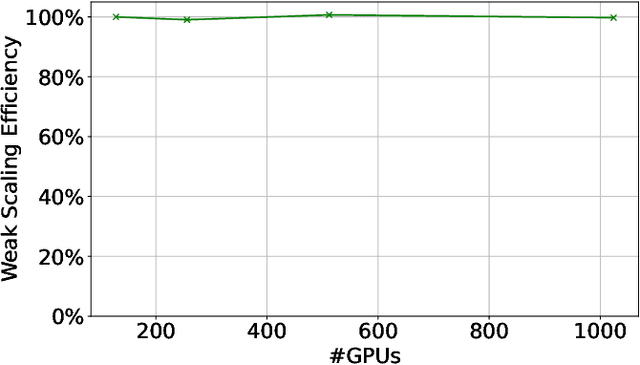
Abstract:Large language models (LLMs) have demonstrated remarkable success as foundational models, benefiting various downstream applications through fine-tuning. Recent studies on loss scaling have demonstrated the superior performance of larger LLMs compared to their smaller counterparts. Nevertheless, training LLMs with billions of parameters poses significant challenges and requires considerable computational resources. For example, training a one trillion parameter GPT-style model on 20 trillion tokens requires a staggering 120 million exaflops of computation. This research explores efficient distributed training strategies to extract this computation from Frontier, the world's first exascale supercomputer dedicated to open science. We enable and investigate various model and data parallel training techniques, such as tensor parallelism, pipeline parallelism, and sharded data parallelism, to facilitate training a trillion-parameter model on Frontier. We empirically assess these techniques and their associated parameters to determine their impact on memory footprint, communication latency, and GPU's computational efficiency. We analyze the complex interplay among these techniques and find a strategy to combine them to achieve high throughput through hyperparameter tuning. We have identified efficient strategies for training large LLMs of varying sizes through empirical analysis and hyperparameter tuning. For 22 Billion, 175 Billion, and 1 Trillion parameters, we achieved GPU throughputs of $38.38\%$, $36.14\%$, and $31.96\%$, respectively. For the training of the 175 Billion parameter model and the 1 Trillion parameter model, we achieved $100\%$ weak scaling efficiency on 1024 and 3072 MI250X GPUs, respectively. We also achieved strong scaling efficiencies of $89\%$ and $87\%$ for these two models.
Ultra-Long Sequence Distributed Transformer
Nov 08, 2023



Abstract:Transformer models trained on long sequences often achieve higher accuracy than short sequences. Unfortunately, conventional transformers struggle with long sequence training due to the overwhelming computation and memory requirements. Existing methods for long sequence training offer limited speedup and memory reduction, and may compromise accuracy. This paper presents a novel and efficient distributed training method, the Long Short-Sequence Transformer (LSS Transformer), for training transformer with long sequences. It distributes a long sequence into segments among GPUs, with each GPU computing a partial self-attention for its segment. Then, it uses a fused communication and a novel double gradient averaging technique to avoid the need to aggregate partial self-attention and minimize communication overhead. We evaluated the performance between LSS Transformer and the state-of-the-art Nvidia sequence parallelism on a Wikipedia enwik8 dataset. Results show that our proposed method lead to 5.6x faster and 10.2x more memory-efficient implementation compared to state-of-the-art sequence parallelism on 144 Nvidia V100 GPUs. Moreover, our algorithm scales to an extreme sequence length of 50,112 at 3,456 GPUs, achieving 161% super-linear parallel efficiency and a throughput of 32 petaflops.
DeepSpeed4Science Initiative: Enabling Large-Scale Scientific Discovery through Sophisticated AI System Technologies
Oct 11, 2023



Abstract:In the upcoming decade, deep learning may revolutionize the natural sciences, enhancing our capacity to model and predict natural occurrences. This could herald a new era of scientific exploration, bringing significant advancements across sectors from drug development to renewable energy. To answer this call, we present DeepSpeed4Science initiative (deepspeed4science.ai) which aims to build unique capabilities through AI system technology innovations to help domain experts to unlock today's biggest science mysteries. By leveraging DeepSpeed's current technology pillars (training, inference and compression) as base technology enablers, DeepSpeed4Science will create a new set of AI system technologies tailored for accelerating scientific discoveries by addressing their unique complexity beyond the common technical approaches used for accelerating generic large language models (LLMs). In this paper, we showcase the early progress we made with DeepSpeed4Science in addressing two of the critical system challenges in structural biology research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge