Mayanka Chandra Shekar
VISION: Toward a Standardized Process for Radiology Image Management at the National Level
Apr 29, 2024Abstract:The compilation and analysis of radiological images poses numerous challenges for researchers. The sheer volume of data as well as the computational needs of algorithms capable of operating on images are extensive. Additionally, the assembly of these images alone is difficult, as these exams may differ widely in terms of clinical context, structured annotation available for model training, modality, and patient identifiers. In this paper, we describe our experiences and challenges in establishing a trusted collection of radiology images linked to the United States Department of Veterans Affairs (VA) electronic health record database. We also discuss implications in making this repository research-ready for medical investigators. Key insights include uncovering the specific procedures required for transferring images from a clinical to a research-ready environment, as well as roadblocks and bottlenecks in this process that may hinder future efforts at automation.
Ultra-Long Sequence Distributed Transformer
Nov 08, 2023



Abstract:Transformer models trained on long sequences often achieve higher accuracy than short sequences. Unfortunately, conventional transformers struggle with long sequence training due to the overwhelming computation and memory requirements. Existing methods for long sequence training offer limited speedup and memory reduction, and may compromise accuracy. This paper presents a novel and efficient distributed training method, the Long Short-Sequence Transformer (LSS Transformer), for training transformer with long sequences. It distributes a long sequence into segments among GPUs, with each GPU computing a partial self-attention for its segment. Then, it uses a fused communication and a novel double gradient averaging technique to avoid the need to aggregate partial self-attention and minimize communication overhead. We evaluated the performance between LSS Transformer and the state-of-the-art Nvidia sequence parallelism on a Wikipedia enwik8 dataset. Results show that our proposed method lead to 5.6x faster and 10.2x more memory-efficient implementation compared to state-of-the-art sequence parallelism on 144 Nvidia V100 GPUs. Moreover, our algorithm scales to an extreme sequence length of 50,112 at 3,456 GPUs, achieving 161% super-linear parallel efficiency and a throughput of 32 petaflops.
Integration of Domain Knowledge using Medical Knowledge Graph Deep Learning for Cancer Phenotyping
Jan 05, 2021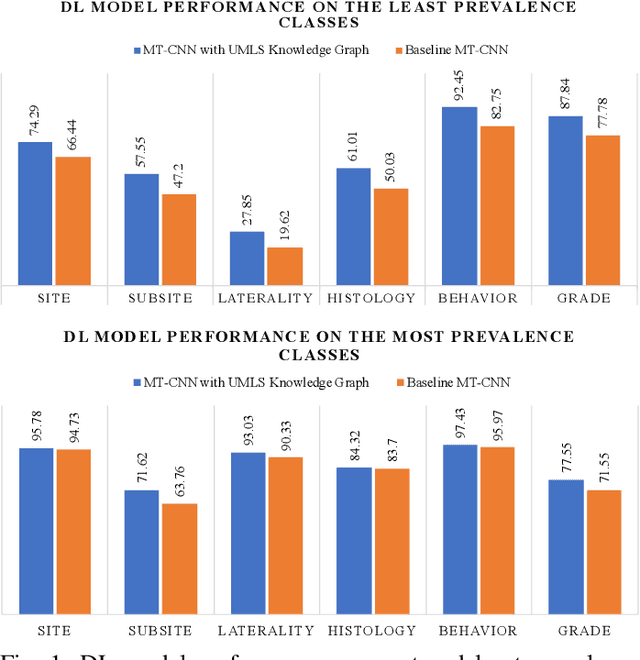
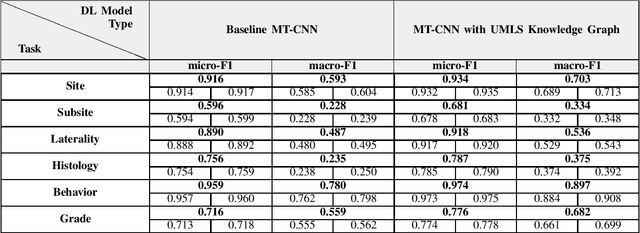
Abstract:A key component of deep learning (DL) for natural language processing (NLP) is word embeddings. Word embeddings that effectively capture the meaning and context of the word that they represent can significantly improve the performance of downstream DL models for various NLP tasks. Many existing word embeddings techniques capture the context of words based on word co-occurrence in documents and text; however, they often cannot capture broader domain-specific relationships between concepts that may be crucial for the NLP task at hand. In this paper, we propose a method to integrate external knowledge from medical terminology ontologies into the context captured by word embeddings. Specifically, we use a medical knowledge graph, such as the unified medical language system (UMLS), to find connections between clinical terms in cancer pathology reports. This approach aims to minimize the distance between connected clinical concepts. We evaluate the proposed approach using a Multitask Convolutional Neural Network (MT-CNN) to extract six cancer characteristics -- site, subsite, laterality, behavior, histology, and grade -- from a dataset of ~900K cancer pathology reports. The results show that the MT-CNN model which uses our domain informed embeddings outperforms the same MT-CNN using standard word2vec embeddings across all tasks, with an improvement in the overall micro- and macro-F1 scores by 4.97\%and 22.5\%, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge