Xiao-Cheng Wu
Louisiana Tumor Registry
Global explainability of a deep abstaining classifier
Apr 01, 2025



Abstract:We present a global explainability method to characterize sources of errors in the histology prediction task of our real-world multitask convolutional neural network (MTCNN)-based deep abstaining classifier (DAC), for automated annotation of cancer pathology reports from NCI-SEER registries. Our classifier was trained and evaluated on 1.04 million hand-annotated samples and makes simultaneous predictions of cancer site, subsite, histology, laterality, and behavior for each report. The DAC framework enables the model to abstain on ambiguous reports and/or confusing classes to achieve a target accuracy on the retained (non-abstained) samples, but at the cost of decreased coverage. Requiring 97% accuracy on the histology task caused our model to retain only 22% of all samples, mostly the less ambiguous and common classes. Local explainability with the GradInp technique provided a computationally efficient way of obtaining contextual reasoning for thousands of individual predictions. Our method, involving dimensionality reduction of approximately 13000 aggregated local explanations, enabled global identification of sources of errors as hierarchical complexity among classes, label noise, insufficient information, and conflicting evidence. This suggests several strategies such as exclusion criteria, focused annotation, and reduced penalties for errors involving hierarchically related classes to iteratively improve our DAC in this complex real-world implementation.
Integration of Domain Knowledge using Medical Knowledge Graph Deep Learning for Cancer Phenotyping
Jan 05, 2021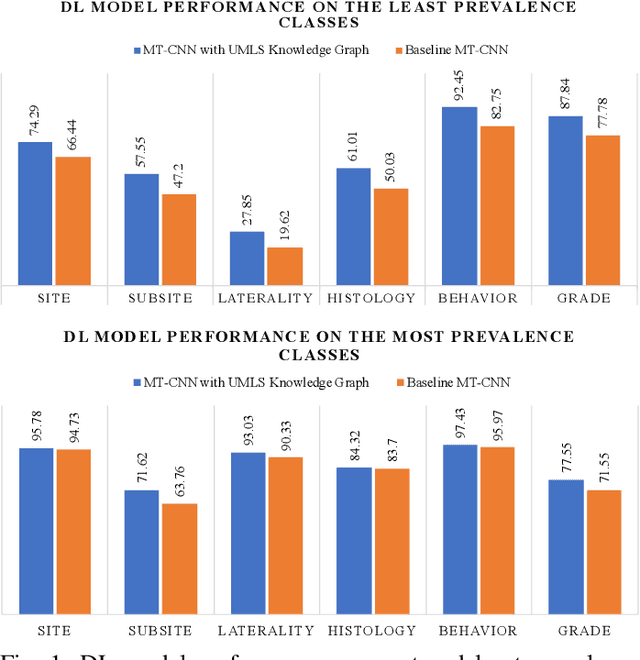
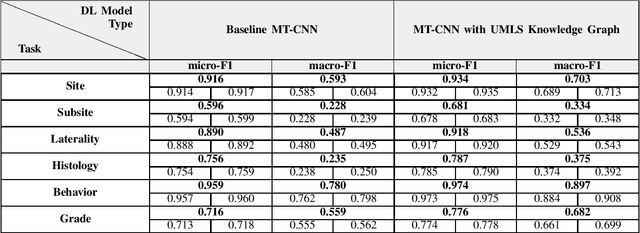
Abstract:A key component of deep learning (DL) for natural language processing (NLP) is word embeddings. Word embeddings that effectively capture the meaning and context of the word that they represent can significantly improve the performance of downstream DL models for various NLP tasks. Many existing word embeddings techniques capture the context of words based on word co-occurrence in documents and text; however, they often cannot capture broader domain-specific relationships between concepts that may be crucial for the NLP task at hand. In this paper, we propose a method to integrate external knowledge from medical terminology ontologies into the context captured by word embeddings. Specifically, we use a medical knowledge graph, such as the unified medical language system (UMLS), to find connections between clinical terms in cancer pathology reports. This approach aims to minimize the distance between connected clinical concepts. We evaluate the proposed approach using a Multitask Convolutional Neural Network (MT-CNN) to extract six cancer characteristics -- site, subsite, laterality, behavior, histology, and grade -- from a dataset of ~900K cancer pathology reports. The results show that the MT-CNN model which uses our domain informed embeddings outperforms the same MT-CNN using standard word2vec embeddings across all tasks, with an improvement in the overall micro- and macro-F1 scores by 4.97\%and 22.5\%, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge