Linda Coyle
Integration of Domain Knowledge using Medical Knowledge Graph Deep Learning for Cancer Phenotyping
Jan 05, 2021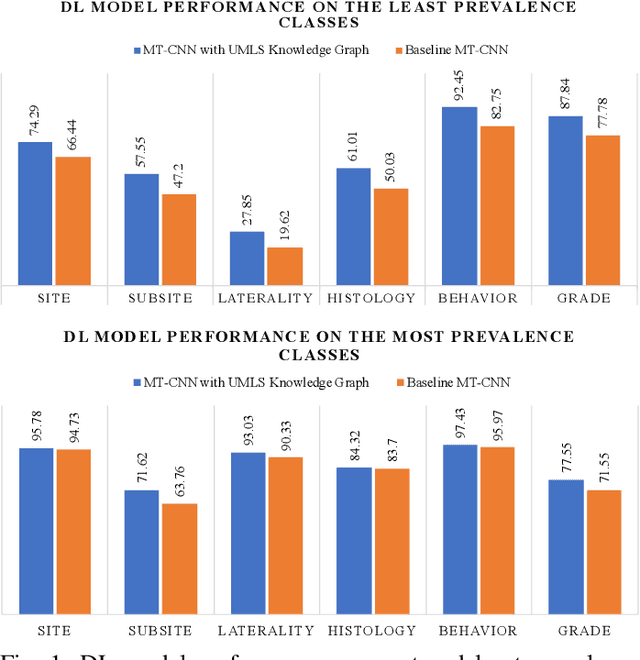
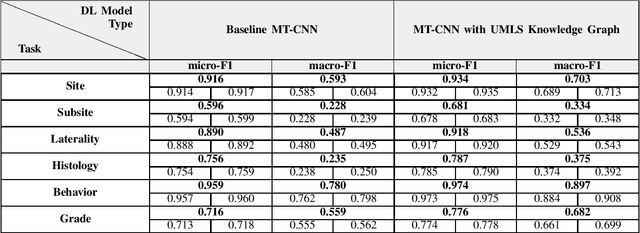
Abstract:A key component of deep learning (DL) for natural language processing (NLP) is word embeddings. Word embeddings that effectively capture the meaning and context of the word that they represent can significantly improve the performance of downstream DL models for various NLP tasks. Many existing word embeddings techniques capture the context of words based on word co-occurrence in documents and text; however, they often cannot capture broader domain-specific relationships between concepts that may be crucial for the NLP task at hand. In this paper, we propose a method to integrate external knowledge from medical terminology ontologies into the context captured by word embeddings. Specifically, we use a medical knowledge graph, such as the unified medical language system (UMLS), to find connections between clinical terms in cancer pathology reports. This approach aims to minimize the distance between connected clinical concepts. We evaluate the proposed approach using a Multitask Convolutional Neural Network (MT-CNN) to extract six cancer characteristics -- site, subsite, laterality, behavior, histology, and grade -- from a dataset of ~900K cancer pathology reports. The results show that the MT-CNN model which uses our domain informed embeddings outperforms the same MT-CNN using standard word2vec embeddings across all tasks, with an improvement in the overall micro- and macro-F1 scores by 4.97\%and 22.5\%, respectively.
Why I'm not Answering: Understanding Determinants of Classification of an Abstaining Classifier for Cancer Pathology Reports
Sep 24, 2020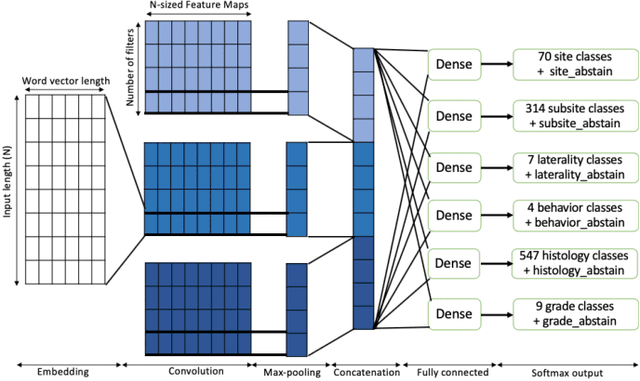
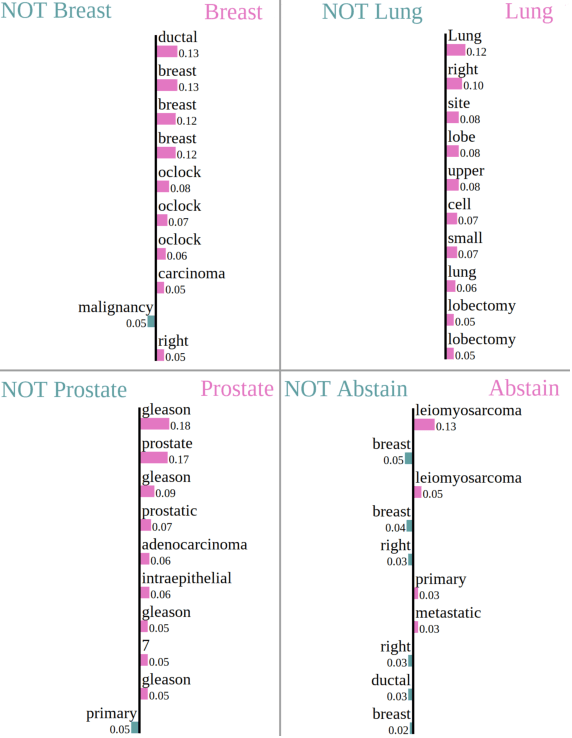
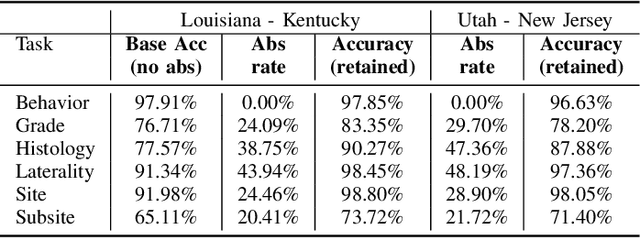
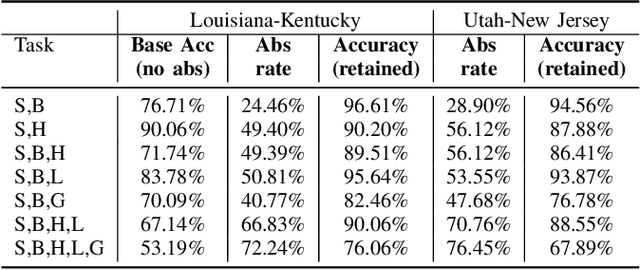
Abstract:Safe deployment of deep learning systems in critical real world applications requires models to make few mistakes, and only under predictable circumstances. Development of such a model is not yet possible, in general. In this work, we address this problem with an abstaining classifier tuned to have $>$95% accuracy, and identify the determinants of abstention with LIME (the Local Interpretable Model-agnostic Explanations method). Essentially, we are training our model to learn the attributes of pathology reports that are likely to lead to incorrect classifications, albeit at the cost of reduced sensitivity. We demonstrate our method in a multitask setting to classify cancer pathology reports from the NCI SEER cancer registries on six tasks of greatest importance. For these tasks, we reduce the classification error rate by factors of 2-5 by abstaining on 25-45% of the reports. For the specific case of cancer site, we are able to identify metastasis and reports involving lymph nodes as responsible for many of the classification mistakes, and that the extent and types of mistakes vary systematically with cancer site (eg. breast, lung, and prostate). When combining across three of the tasks, our model classifies 50% of the reports with an accuracy greater than 95% for three of the six tasks and greater than 85% for all six tasks on the retained samples. By using this information, we expect to define work flows that incorporate machine learning only in the areas where it is sufficiently robust and accurate, saving human attention to areas where it is required.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge