Kumkum Ganguly
Los Alamos National Laboratory
Global explainability of a deep abstaining classifier
Apr 01, 2025



Abstract:We present a global explainability method to characterize sources of errors in the histology prediction task of our real-world multitask convolutional neural network (MTCNN)-based deep abstaining classifier (DAC), for automated annotation of cancer pathology reports from NCI-SEER registries. Our classifier was trained and evaluated on 1.04 million hand-annotated samples and makes simultaneous predictions of cancer site, subsite, histology, laterality, and behavior for each report. The DAC framework enables the model to abstain on ambiguous reports and/or confusing classes to achieve a target accuracy on the retained (non-abstained) samples, but at the cost of decreased coverage. Requiring 97% accuracy on the histology task caused our model to retain only 22% of all samples, mostly the less ambiguous and common classes. Local explainability with the GradInp technique provided a computationally efficient way of obtaining contextual reasoning for thousands of individual predictions. Our method, involving dimensionality reduction of approximately 13000 aggregated local explanations, enabled global identification of sources of errors as hierarchical complexity among classes, label noise, insufficient information, and conflicting evidence. This suggests several strategies such as exclusion criteria, focused annotation, and reduced penalties for errors involving hierarchically related classes to iteratively improve our DAC in this complex real-world implementation.
Why I'm not Answering: Understanding Determinants of Classification of an Abstaining Classifier for Cancer Pathology Reports
Sep 24, 2020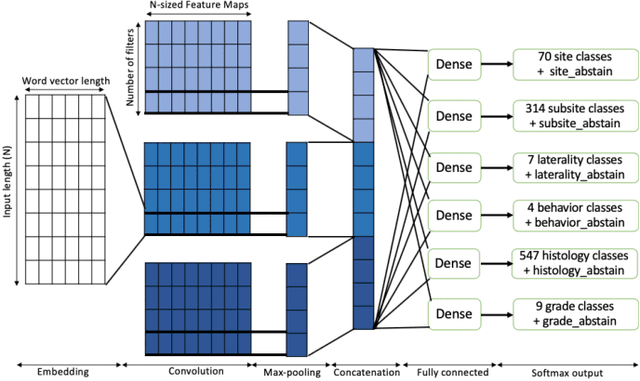
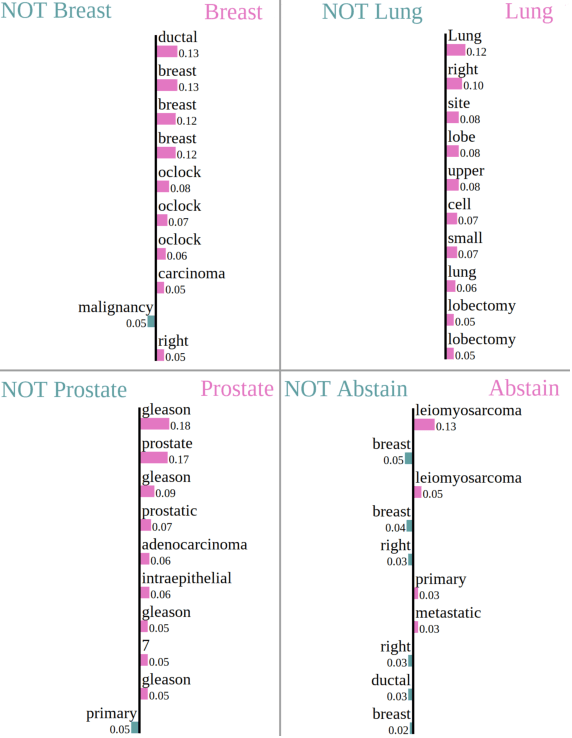
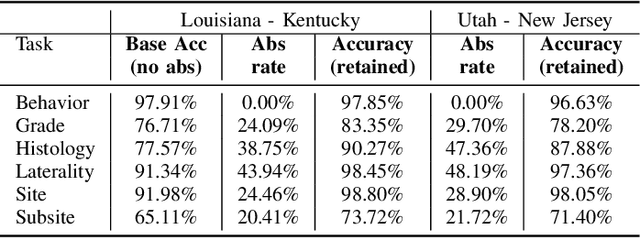
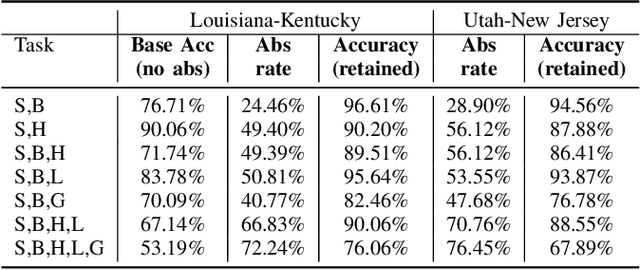
Abstract:Safe deployment of deep learning systems in critical real world applications requires models to make few mistakes, and only under predictable circumstances. Development of such a model is not yet possible, in general. In this work, we address this problem with an abstaining classifier tuned to have $>$95% accuracy, and identify the determinants of abstention with LIME (the Local Interpretable Model-agnostic Explanations method). Essentially, we are training our model to learn the attributes of pathology reports that are likely to lead to incorrect classifications, albeit at the cost of reduced sensitivity. We demonstrate our method in a multitask setting to classify cancer pathology reports from the NCI SEER cancer registries on six tasks of greatest importance. For these tasks, we reduce the classification error rate by factors of 2-5 by abstaining on 25-45% of the reports. For the specific case of cancer site, we are able to identify metastasis and reports involving lymph nodes as responsible for many of the classification mistakes, and that the extent and types of mistakes vary systematically with cancer site (eg. breast, lung, and prostate). When combining across three of the tasks, our model classifies 50% of the reports with an accuracy greater than 95% for three of the six tasks and greater than 85% for all six tasks on the retained samples. By using this information, we expect to define work flows that incorporate machine learning only in the areas where it is sufficiently robust and accurate, saving human attention to areas where it is required.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge