Dong Sun
ERNIE 5.0 Technical Report
Feb 04, 2026Abstract:In this report, we introduce ERNIE 5.0, a natively autoregressive foundation model desinged for unified multimodal understanding and generation across text, image, video, and audio. All modalities are trained from scratch under a unified next-group-of-tokens prediction objective, based on an ultra-sparse mixture-of-experts (MoE) architecture with modality-agnostic expert routing. To address practical challenges in large-scale deployment under diverse resource constraints, ERNIE 5.0 adopts a novel elastic training paradigm. Within a single pre-training run, the model learns a family of sub-models with varying depths, expert capacities, and routing sparsity, enabling flexible trade-offs among performance, model size, and inference latency in memory- or time-constrained scenarios. Moreover, we systematically address the challenges of scaling reinforcement learning to unified foundation models, thereby guaranteeing efficient and stable post-training under ultra-sparse MoE architectures and diverse multimodal settings. Extensive experiments demonstrate that ERNIE 5.0 achieves strong and balanced performance across multiple modalities. To the best of our knowledge, among publicly disclosed models, ERNIE 5.0 represents the first production-scale realization of a trillion-parameter unified autoregressive model that supports both multimodal understanding and generation. To facilitate further research, we present detailed visualizations of modality-agnostic expert routing in the unified model, alongside comprehensive empirical analysis of elastic training, aiming to offer profound insights to the community.
Robustness of Mixtures of Experts to Feature Noise
Jan 21, 2026Abstract:Despite their practical success, it remains unclear why Mixture of Experts (MoE) models can outperform dense networks beyond sheer parameter scaling. We study an iso-parameter regime where inputs exhibit latent modular structure but are corrupted by feature noise, a proxy for noisy internal activations. We show that sparse expert activation acts as a noise filter: compared to a dense estimator, MoEs achieve lower generalization error under feature noise, improved robustness to perturbations, and faster convergence speed. Empirical results on synthetic data and real-world language tasks corroborate the theoretical insights, demonstrating consistent robustness and efficiency gains from sparse modular computation.
Contextual Embedding Learning to Enhance 2D Networks for Volumetric Image Segmentation
Apr 02, 2024Abstract:The segmentation of organs in volumetric medical images plays an important role in computer-aided diagnosis and treatment/surgery planning. Conventional 2D convolutional neural networks (CNNs) can hardly exploit the spatial correlation of volumetric data. Current 3D CNNs have the advantage to extract more powerful volumetric representations but they usually suffer from occupying excessive memory and computation nevertheless. In this study we aim to enhance the 2D networks with contextual information for better volumetric image segmentation. Accordingly, we propose a contextual embedding learning approach to facilitate 2D CNNs capturing spatial information properly. Our approach leverages the learned embedding and the slice-wisely neighboring matching as a soft cue to guide the network. In such a way, the contextual information can be transferred slice-by-slice thus boosting the volumetric representation of the network. Experiments on challenging prostate MRI dataset (PROMISE12) and abdominal CT dataset (CHAOS) show that our contextual embedding learning can effectively leverage the inter-slice context and improve segmentation performance. The proposed approach is a plug-and-play, and memory-efficient solution to enhance the 2D networks for volumetric segmentation. The code will be publicly available.
STOPNet: Multiview-based 6-DoF Suction Detection for Transparent Objects on Production Lines
Oct 09, 2023



Abstract:In this work, we present STOPNet, a framework for 6-DoF object suction detection on production lines, with a focus on but not limited to transparent objects, which is an important and challenging problem in robotic systems and modern industry. Current methods requiring depth input fail on transparent objects due to depth cameras' deficiency in sensing their geometry, while we proposed a novel framework to reconstruct the scene on the production line depending only on RGB input, based on multiview stereo. Compared to existing works, our method not only reconstructs the whole 3D scene in order to obtain high-quality 6-DoF suction poses in real time but also generalizes to novel environments, novel arrangements and novel objects, including challenging transparent objects, both in simulation and the real world. Extensive experiments in simulation and the real world show that our method significantly surpasses the baselines and has better generalizability, which caters to practical industrial needs.
Autonomous Intelligent Navigation for Flexible Endoscopy Using Monocular Depth Guidance and 3-D Shape Planning
Feb 26, 2023



Abstract:Recent advancements toward perception and decision-making of flexible endoscopes have shown great potential in computer-aided surgical interventions. However, owing to modeling uncertainty and inter-patient anatomical variation in flexible endoscopy, the challenge remains for efficient and safe navigation in patient-specific scenarios. This paper presents a novel data-driven framework with self-contained visual-shape fusion for autonomous intelligent navigation of flexible endoscopes requiring no priori knowledge of system models and global environments. A learning-based adaptive visual servoing controller is proposed to online update the eye-in-hand vision-motor configuration and steer the endoscope, which is guided by monocular depth estimation via a vision transformer (ViT). To prevent unnecessary and excessive interactions with surrounding anatomy, an energy-motivated shape planning algorithm is introduced through entire endoscope 3-D proprioception from embedded fiber Bragg grating (FBG) sensors. Furthermore, a model predictive control (MPC) strategy is developed to minimize the elastic potential energy flow and simultaneously optimize the steering policy. Dedicated navigation experiments on a robotic-assisted flexible endoscope with an FBG fiber in several phantom environments demonstrate the effectiveness and adaptability of the proposed framework.
Dual-View Selective Instance Segmentation Network for Unstained Live Adherent Cells in Differential Interference Contrast Images
Jan 27, 2023



Abstract:Despite recent advances in data-independent and deep-learning algorithms, unstained live adherent cell instance segmentation remains a long-standing challenge in cell image processing. Adherent cells' inherent visual characteristics, such as low contrast structures, fading edges, and irregular morphology, have made it difficult to distinguish from one another, even by human experts, let alone computational methods. In this study, we developed a novel deep-learning algorithm called dual-view selective instance segmentation network (DVSISN) for segmenting unstained adherent cells in differential interference contrast (DIC) images. First, we used a dual-view segmentation (DVS) method with pairs of original and rotated images to predict the bounding box and its corresponding mask for each cell instance. Second, we used a mask selection (MS) method to filter the cell instances predicted by the DVS to keep masks closest to the ground truth only. The developed algorithm was trained and validated on our dataset containing 520 images and 12198 cells. Experimental results demonstrate that our algorithm achieves an AP_segm of 0.555, which remarkably overtakes a benchmark by a margin of 23.6%. This study's success opens up a new possibility of using rotated images as input for better prediction in cell images.
Distilled Visual and Robot Kinematics Embeddings for Metric Depth Estimation in Monocular Scene Reconstruction
Nov 27, 2022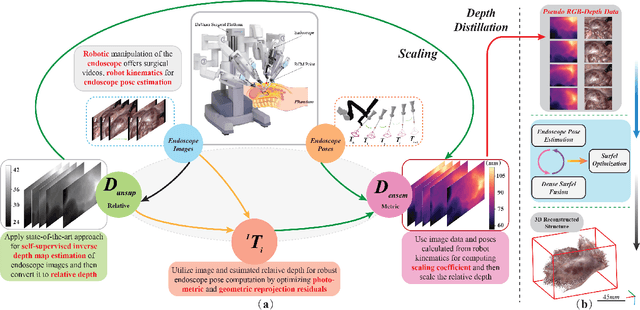
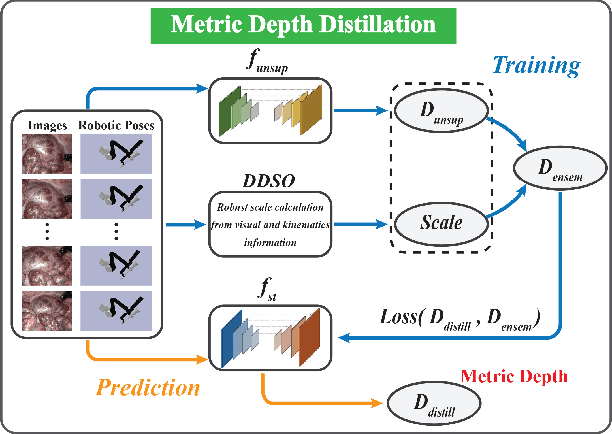
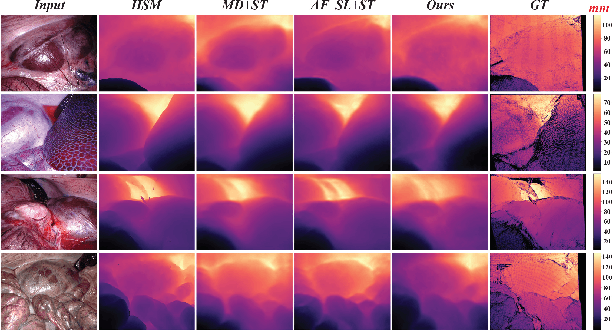
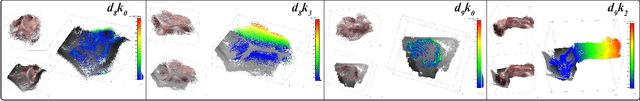
Abstract:Estimating precise metric depth and scene reconstruction from monocular endoscopy is a fundamental task for surgical navigation in robotic surgery. However, traditional stereo matching adopts binocular images to perceive the depth information, which is difficult to transfer to the soft robotics-based surgical systems due to the use of monocular endoscopy. In this paper, we present a novel framework that combines robot kinematics and monocular endoscope images with deep unsupervised learning into a single network for metric depth estimation and then achieve 3D reconstruction of complex anatomy. Specifically, we first obtain the relative depth maps of surgical scenes by leveraging a brightness-aware monocular depth estimation method. Then, the corresponding endoscope poses are computed based on non-linear optimization of geometric and photometric reprojection residuals. Afterwards, we develop a Depth-driven Sliding Optimization (DDSO) algorithm to extract the scaling coefficient from kinematics and calculated poses offline. By coupling the metric scale and relative depth data, we form a robust ensemble that represents the metric and consistent depth. Next, we treat the ensemble as supervisory labels to train a metric depth estimation network for surgeries (i.e., MetricDepthS-Net) that distills the embeddings from the robot kinematics, endoscopic videos, and poses. With accurate metric depth estimation, we utilize a dense visual reconstruction method to recover the 3D structure of the whole surgical site. We have extensively evaluated the proposed framework on public SCARED and achieved comparable performance with stereo-based depth estimation methods. Our results demonstrate the feasibility of the proposed approach to recover the metric depth and 3D structure with monocular inputs.
Stereo Dense Scene Reconstruction and Accurate Laparoscope Localization for Learning-Based Navigation in Robot-Assisted Surgery
Oct 08, 2021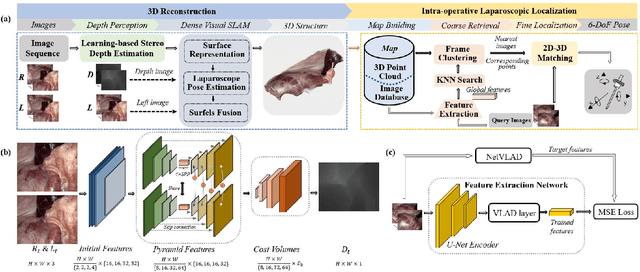
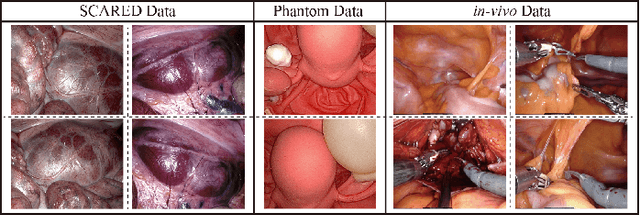
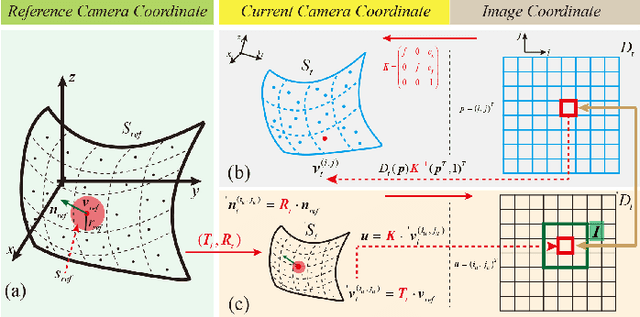
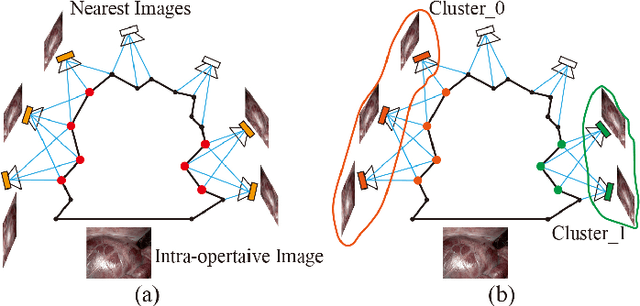
Abstract:The computation of anatomical information and laparoscope position is a fundamental block of robot-assisted surgical navigation in Minimally Invasive Surgery (MIS). Recovering a dense 3D structure of surgical scene using visual cues remains a challenge, and the online laparoscopic tracking mostly relies on external sensors, which increases system complexity. In this paper, we propose a learning-driven framework, in which an image-guided laparoscopic localization with 3D reconstructions of complex anatomical structures is hereby achieved. To reconstruct the 3D structure of the whole surgical environment, we first fine-tune a learning-based stereoscopic depth perception method, which is robust to the texture-less and variant soft tissues, for depth estimation. Then, we develop a dense visual reconstruction algorithm to represent the scene by surfels, estimate the laparoscope pose and fuse the depth data into a unified reference coordinate for tissue reconstruction. To estimate poses of new laparoscope views, we realize a coarse-to-fine localization method, which incorporates our reconstructed 3D model. We evaluate the reconstruction method and the localization module on three datasets, namely, the stereo correspondence and reconstruction of endoscopic data (SCARED), the ex-vivo phantom and tissue data collected with Universal Robot (UR) and Karl Storz Laparoscope, and the in-vivo DaVinci robotic surgery dataset. Extensive experiments have been conducted to prove the superior performance of our method in 3D anatomy reconstruction and laparoscopic localization, which demonstrates its potential implementation to surgical navigation system.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge