Lian Ding
STOPNet: Multiview-based 6-DoF Suction Detection for Transparent Objects on Production Lines
Oct 09, 2023



Abstract:In this work, we present STOPNet, a framework for 6-DoF object suction detection on production lines, with a focus on but not limited to transparent objects, which is an important and challenging problem in robotic systems and modern industry. Current methods requiring depth input fail on transparent objects due to depth cameras' deficiency in sensing their geometry, while we proposed a novel framework to reconstruct the scene on the production line depending only on RGB input, based on multiview stereo. Compared to existing works, our method not only reconstructs the whole 3D scene in order to obtain high-quality 6-DoF suction poses in real time but also generalizes to novel environments, novel arrangements and novel objects, including challenging transparent objects, both in simulation and the real world. Extensive experiments in simulation and the real world show that our method significantly surpasses the baselines and has better generalizability, which caters to practical industrial needs.
Intrinsic Bias Identification on Medical Image Datasets
Mar 29, 2022



Abstract:Machine learning based medical image analysis highly depends on datasets. Biases in the dataset can be learned by the model and degrade the generalizability of the applications. There are studies on debiased models. However, scientists and practitioners are difficult to identify implicit biases in the datasets, which causes lack of reliable unbias test datasets to valid models. To tackle this issue, we first define the data intrinsic bias attribute, and then propose a novel bias identification framework for medical image datasets. The framework contains two major components, KlotskiNet and Bias Discriminant Direction Analysis(bdda), where KlostkiNet is to build the mapping which makes backgrounds to distinguish positive and negative samples and bdda provides a theoretical solution on determining bias attributes. Experimental results on three datasets show the effectiveness of the bias attributes discovered by the framework.
Learning Incrementally to Segment Multiple Organs in a CT Image
Mar 04, 2022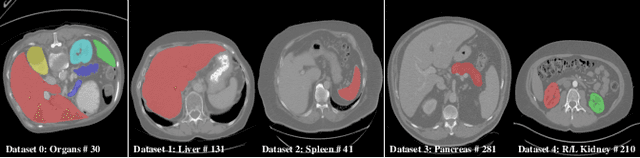
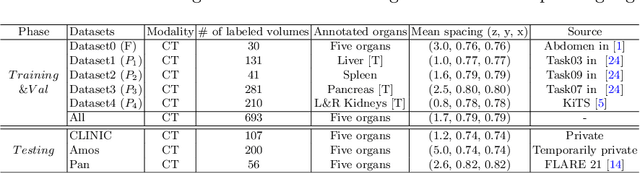
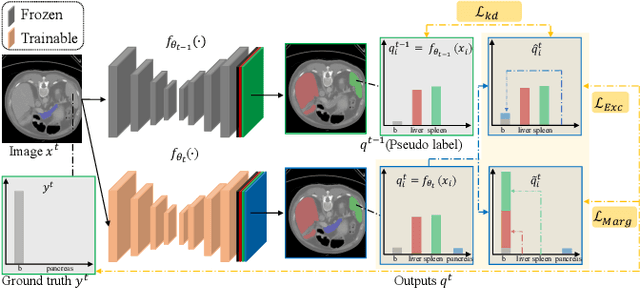
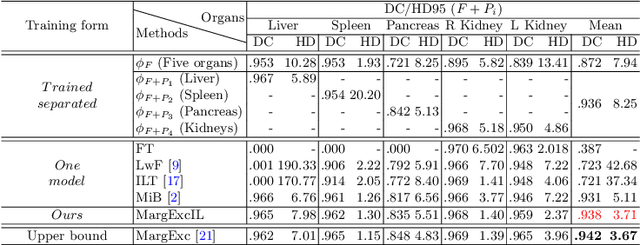
Abstract:There exists a large number of datasets for organ segmentation, which are partially annotated and sequentially constructed. A typical dataset is constructed at a certain time by curating medical images and annotating the organs of interest. In other words, new datasets with annotations of new organ categories are built over time. To unleash the potential behind these partially labeled, sequentially-constructed datasets, we propose to incrementally learn a multi-organ segmentation model. In each incremental learning (IL) stage, we lose the access to previous data and annotations, whose knowledge is assumingly captured by the current model, and gain the access to a new dataset with annotations of new organ categories, from which we learn to update the organ segmentation model to include the new organs. While IL is notorious for its `catastrophic forgetting' weakness in the context of natural image analysis, we experimentally discover that such a weakness mostly disappears for CT multi-organ segmentation. To further stabilize the model performance across the IL stages, we introduce a light memory module and some loss functions to restrain the representation of different categories in feature space, aggregating feature representation of the same class and separating feature representation of different classes. Extensive experiments on five open-sourced datasets are conducted to illustrate the effectiveness of our method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge