Declan P. O'Regan
Inference-Time Dynamic Modality Selection for Incomplete Multimodal Classification
Jan 30, 2026Abstract:Multimodal deep learning (MDL) has achieved remarkable success across various domains, yet its practical deployment is often hindered by incomplete multimodal data. Existing incomplete MDL methods either discard missing modalities, risking the loss of valuable task-relevant information, or recover them, potentially introducing irrelevant noise, leading to the discarding-imputation dilemma. To address this dilemma, in this paper, we propose DyMo, a new inference-time dynamic modality selection framework that adaptively identifies and integrates reliable recovered modalities, fully exploring task-relevant information beyond the conventional discard-or-impute paradigm. Central to DyMo is a novel selection algorithm that maximizes multimodal task-relevant information for each test sample. Since direct estimation of such information at test time is intractable due to the unknown data distribution, we theoretically establish a connection between information and the task loss, which we compute at inference time as a tractable proxy. Building on this, a novel principled reward function is proposed to guide modality selection. In addition, we design a flexible multimodal network architecture compatible with arbitrary modality combinations, alongside a tailored training strategy for robust representation learning. Extensive experiments on diverse natural and medical image datasets show that DyMo significantly outperforms state-of-the-art incomplete/dynamic MDL methods across various missing-data scenarios. Our code is available at https://github.com//siyi-wind/DyMo.
Multimodal Conditional MeshGAN for Personalized Aneurysm Growth Prediction
Aug 27, 2025Abstract:Personalized, accurate prediction of aortic aneurysm progression is essential for timely intervention but remains challenging due to the need to model both subtle local deformations and global anatomical changes within complex 3D geometries. We propose MCMeshGAN, the first multimodal conditional mesh-to-mesh generative adversarial network for 3D aneurysm growth prediction. MCMeshGAN introduces a dual-branch architecture combining a novel local KNN-based convolutional network (KCN) to preserve fine-grained geometric details and a global graph convolutional network (GCN) to capture long-range structural context, overcoming the over-smoothing limitations of deep GCNs. A dedicated condition branch encodes clinical attributes (age, sex) and the target time interval to generate anatomically plausible, temporally controlled predictions, enabling retrospective and prospective modeling. We curated TAAMesh, a new longitudinal thoracic aortic aneurysm mesh dataset consisting of 590 multimodal records (CT scans, 3D meshes, and clinical data) from 208 patients. Extensive experiments demonstrate that MCMeshGAN consistently outperforms state-of-the-art baselines in both geometric accuracy and clinically important diameter estimation. This framework offers a robust step toward clinically deployable, personalized 3D disease trajectory modeling. The source code for MCMeshGAN and the baseline methods is publicly available at https://github.com/ImperialCollegeLondon/MCMeshGAN.
STiL: Semi-supervised Tabular-Image Learning for Comprehensive Task-Relevant Information Exploration in Multimodal Classification
Mar 08, 2025Abstract:Multimodal image-tabular learning is gaining attention, yet it faces challenges due to limited labeled data. While earlier work has applied self-supervised learning (SSL) to unlabeled data, its task-agnostic nature often results in learning suboptimal features for downstream tasks. Semi-supervised learning (SemiSL), which combines labeled and unlabeled data, offers a promising solution. However, existing multimodal SemiSL methods typically focus on unimodal or modality-shared features, ignoring valuable task-relevant modality-specific information, leading to a Modality Information Gap. In this paper, we propose STiL, a novel SemiSL tabular-image framework that addresses this gap by comprehensively exploring task-relevant information. STiL features a new disentangled contrastive consistency module to learn cross-modal invariant representations of shared information while retaining modality-specific information via disentanglement. We also propose a novel consensus-guided pseudo-labeling strategy to generate reliable pseudo-labels based on classifier consensus, along with a new prototype-guided label smoothing technique to refine pseudo-label quality with prototype embeddings, thereby enhancing task-relevant information learning in unlabeled data. Experiments on natural and medical image datasets show that STiL outperforms the state-of-the-art supervised/SSL/SemiSL image/multimodal approaches. Our code is publicly available.
TIP: Tabular-Image Pre-training for Multimodal Classification with Incomplete Data
Jul 10, 2024



Abstract:Images and structured tables are essential parts of real-world databases. Though tabular-image representation learning is promising to create new insights, it remains a challenging task, as tabular data is typically heterogeneous and incomplete, presenting significant modality disparities with images. Earlier works have mainly focused on simple modality fusion strategies in complete data scenarios, without considering the missing data issue, and thus are limited in practice. In this paper, we propose TIP, a novel tabular-image pre-training framework for learning multimodal representations robust to incomplete tabular data. Specifically, TIP investigates a novel self-supervised learning (SSL) strategy, including a masked tabular reconstruction task for tackling data missingness, and image-tabular matching and contrastive learning objectives to capture multimodal information. Moreover, TIP proposes a versatile tabular encoder tailored for incomplete, heterogeneous tabular data and a multimodal interaction module for inter-modality representation learning. Experiments are performed on downstream multimodal classification tasks using both natural and medical image datasets. The results show that TIP outperforms state-of-the-art supervised/SSL image/multimodal algorithms in both complete and incomplete data scenarios. Our code is available at https://github.com/siyi-wind/TIP.
CHeart: A Conditional Spatio-Temporal Generative Model for Cardiac Anatomy
Jan 30, 2023Abstract:Two key questions in cardiac image analysis are to assess the anatomy and motion of the heart from images; and to understand how they are associated with non-imaging clinical factors such as gender, age and diseases. While the first question can often be addressed by image segmentation and motion tracking algorithms, our capability to model and to answer the second question is still limited. In this work, we propose a novel conditional generative model to describe the 4D spatio-temporal anatomy of the heart and its interaction with non-imaging clinical factors. The clinical factors are integrated as the conditions of the generative modelling, which allows us to investigate how these factors influence the cardiac anatomy. We evaluate the model performance in mainly two tasks, anatomical sequence completion and sequence generation. The model achieves a high performance in anatomical sequence completion, comparable to or outperforming other state-of-the-art generative models. In terms of sequence generation, given clinical conditions, the model can generate realistic synthetic 4D sequential anatomies that share similar distributions with the real data.
Data-driven generation of 4D velocity profiles in the aneurysmal ascending aorta
Nov 01, 2022Abstract:Numerical simulations of blood flow are a valuable tool to investigate the pathophysiology of ascending thoracic aortic aneurysms (ATAA). To accurately reproduce hemodynamics, computational fluid dynamics (CFD) models must employ realistic inflow boundary conditions (BCs). However, the limited availability of in vivo velocity measurements still makes researchers resort to idealized BCs. In this study we generated and thoroughly characterized a large dataset of synthetic 4D aortic velocity profiles suitable to be used as BCs for CFD simulations. 4D flow MRI scans of 30 subjects with ATAA were processed to extract cross-sectional planes along the ascending aorta, ensuring spatial alignment among all planes and interpolating all velocity fields to a reference configuration. Velocity profiles of the clinical cohort were extensively characterized by computing flow morphology descriptors of both spatial and temporal features. By exploiting principal component analysis (PCA), a statistical shape model (SSM) of 4D aortic velocity profiles was built and a dataset of 437 synthetic cases with realistic properties was generated. Comparison between clinical and synthetic datasets showed that the synthetic data presented similar characteristics as the clinical population in terms of key morphological parameters. The average velocity profile qualitatively resembled a parabolic-shaped profile, but was quantitatively characterized by more complex flow patterns which an idealized profile would not replicate. Statistically significant correlations were found between PCA principal modes of variation and flow descriptors. We built a data-driven generative model of 4D aortic velocity profiles, suitable to be used in computational studies of blood flow. The proposed software system also allows to map any of the generated velocity profiles to the inlet plane of any virtual subject given its coordinate set.
Nesterov Accelerated ADMM for Fast Diffeomorphic Image Registration
Sep 26, 2021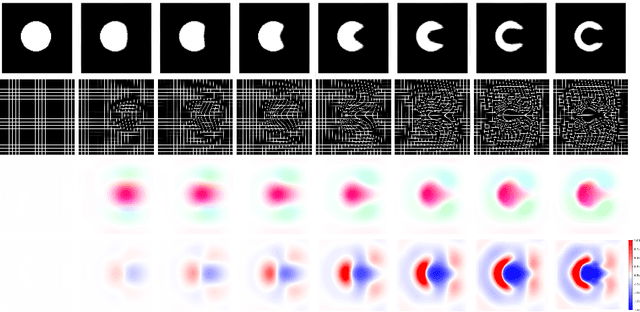
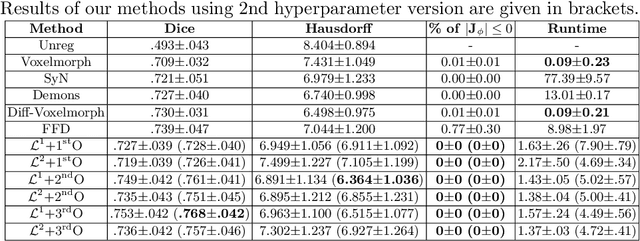
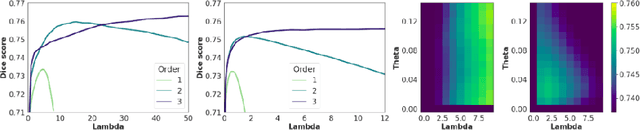
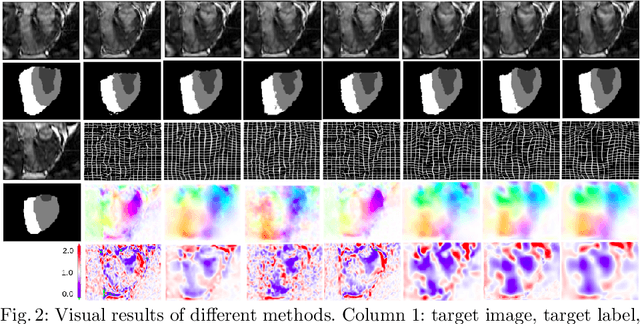
Abstract:Deterministic approaches using iterative optimisation have been historically successful in diffeomorphic image registration (DiffIR). Although these approaches are highly accurate, they typically carry a significant computational burden. Recent developments in stochastic approaches based on deep learning have achieved sub-second runtimes for DiffIR with competitive registration accuracy, offering a fast alternative to conventional iterative methods. In this paper, we attempt to reduce this difference in speed whilst retaining the performance advantage of iterative approaches in DiffIR. We first propose a simple iterative scheme that functionally composes intermediate non-stationary velocity fields to handle large deformations in images whilst guaranteeing diffeomorphisms in the resultant deformation. We then propose a convex optimisation model that uses a regularisation term of arbitrary order to impose smoothness on these velocity fields and solve this model with a fast algorithm that combines Nesterov gradient descent and the alternating direction method of multipliers (ADMM). Finally, we leverage the computational power of GPU to implement this accelerated ADMM solver on a 3D cardiac MRI dataset, further reducing runtime to less than 2 seconds. In addition to producing strictly diffeomorphic deformations, our methods outperform both state-of-the-art deep learning-based and iterative DiffIR approaches in terms of dice and Hausdorff scores, with speed approaching the inference time of deep learning-based methods.
Joint Semi-supervised 3D Super-Resolution and Segmentation with Mixed Adversarial Gaussian Domain Adaptation
Jul 16, 2021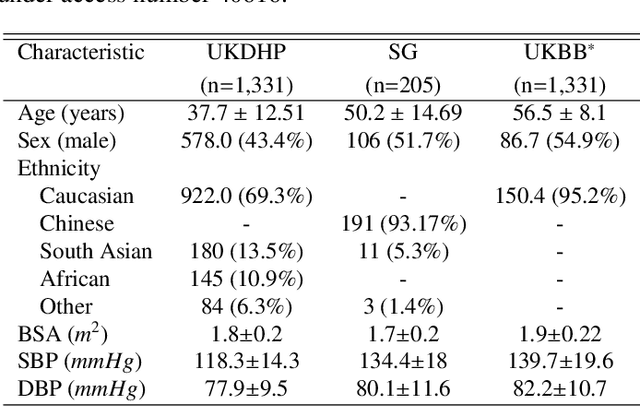
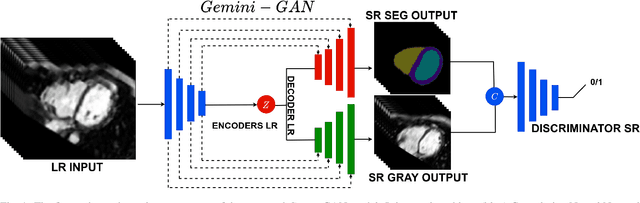
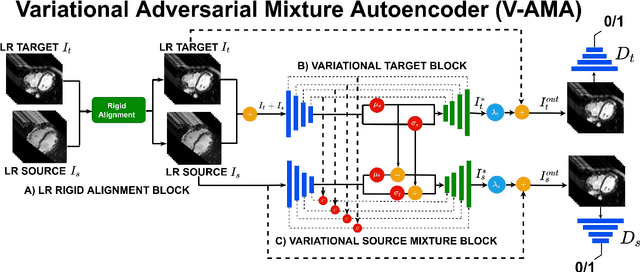
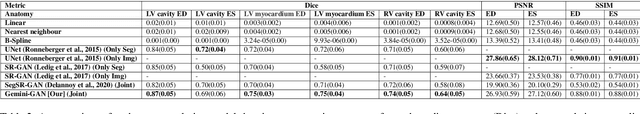
Abstract:Optimising the analysis of cardiac structure and function requires accurate 3D representations of shape and motion. However, techniques such as cardiac magnetic resonance imaging are conventionally limited to acquiring contiguous cross-sectional slices with low through-plane resolution and potential inter-slice spatial misalignment. Super-resolution in medical imaging aims to increase the resolution of images but is conventionally trained on features from low resolution datasets and does not super-resolve corresponding segmentations. Here we propose a semi-supervised multi-task generative adversarial network (Gemini-GAN) that performs joint super-resolution of the images and their labels using a ground truth of high resolution 3D cines and segmentations, while an unsupervised variational adversarial mixture autoencoder (V-AMA) is used for continuous domain adaptation. Our proposed approach is extensively evaluated on two transnational multi-ethnic populations of 1,331 and 205 adults respectively, delivering an improvement on state of the art methods in terms of Dice index, peak signal to noise ratio, and structural similarity index measure. This framework also exceeds the performance of state of the art generative domain adaptation models on external validation (Dice index 0.81 vs 0.74 for the left ventricle). This demonstrates how joint super-resolution and segmentation, trained on 3D ground-truth data with cross-domain generalization, enables robust precision phenotyping in diverse populations.
Learning a Model-Driven Variational Network for Deformable Image Registration
May 25, 2021



Abstract:Data-driven deep learning approaches to image registration can be less accurate than conventional iterative approaches, especially when training data is limited. To address this whilst retaining the fast inference speed of deep learning, we propose VR-Net, a novel cascaded variational network for unsupervised deformable image registration. Using the variable splitting optimization scheme, we first convert the image registration problem, established in a generic variational framework, into two sub-problems, one with a point-wise, closed-form solution while the other one is a denoising problem. We then propose two neural layers (i.e. warping layer and intensity consistency layer) to model the analytical solution and a residual U-Net to formulate the denoising problem (i.e. generalized denoising layer). Finally, we cascade the warping layer, intensity consistency layer, and generalized denoising layer to form the VR-Net. Extensive experiments on three (two 2D and one 3D) cardiac magnetic resonance imaging datasets show that VR-Net outperforms state-of-the-art deep learning methods on registration accuracy, while maintains the fast inference speed of deep learning and the data-efficiency of variational model.
Explainable Shape Analysis through Deep Hierarchical Generative Models: Application to Cardiac Remodeling
Jun 28, 2019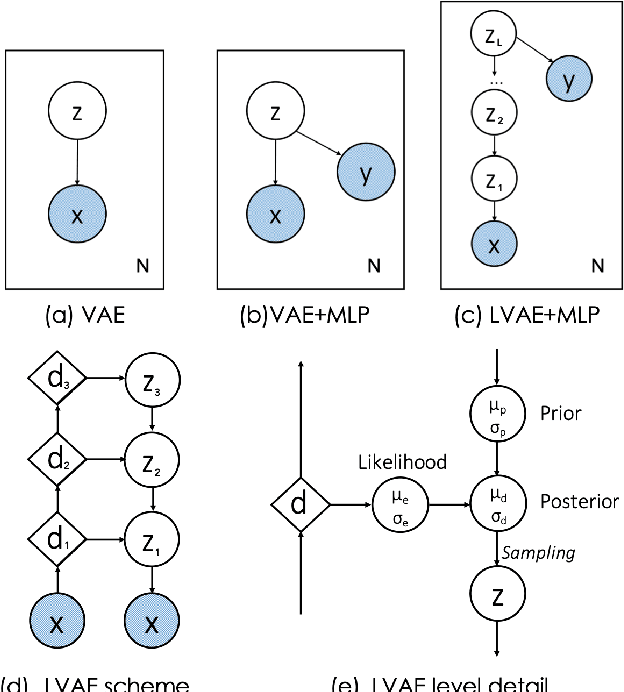
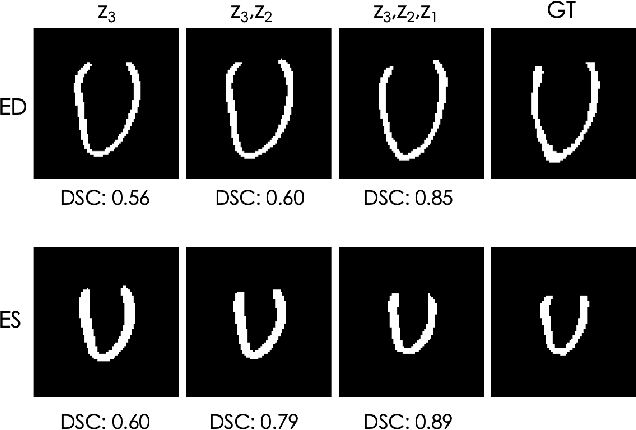
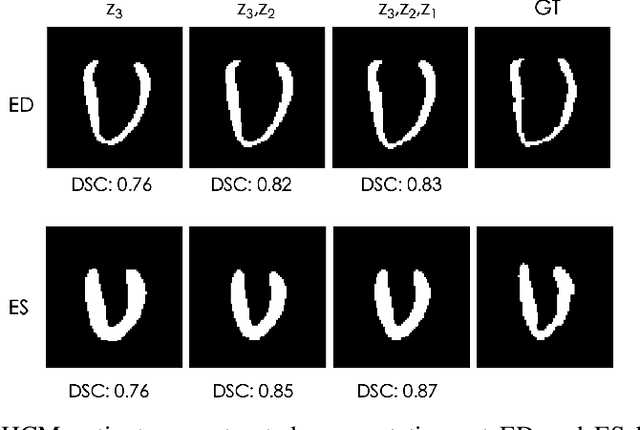
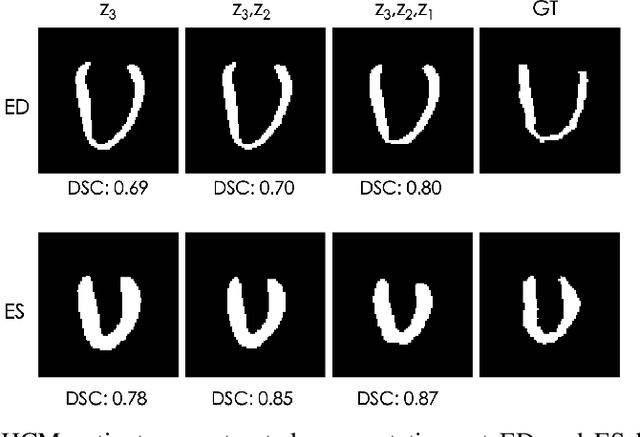
Abstract:Quantification of anatomical shape changes still relies on scalar global indexes which are largely insensitive to regional or asymmetric modifications. Accurate assessment of pathology-driven anatomical remodeling is a crucial step for the diagnosis and treatment of heart conditions. Deep learning approaches have recently achieved wide success in the analysis of medical images, but they lack interpretability in the feature extraction and decision processes. In this work, we propose a new interpretable deep learning model for shape analysis. In particular, we exploit deep generative networks to model a population of anatomical segmentations through a hierarchy of conditional latent variables. At the highest level of this hierarchy, a two-dimensional latent space is simultaneously optimised to discriminate distinct clinical conditions, enabling the direct visualisation of the classification space. Moreover, the anatomical variability encoded by this discriminative latent space can be visualised in the segmentation space thanks to the generative properties of the model, making the classification task transparent. This approach yielded high accuracy in the categorisation of healthy and remodelled hearts when tested on unseen segmentations from our own multi-centre dataset as well as in an external validation set. More importantly, it enabled the visualisation in three-dimensions of the most discriminative anatomical features between the two conditions. The proposed approach scales effectively to large populations, facilitating high-throughput analysis of normal anatomy and pathology in large-scale studies of volumetric imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge