Buyue Qian
MixDec Sampling: A Soft Link-based Sampling Method of Graph Neural Network for Recommendation
Feb 12, 2025Abstract:Graph neural networks have been widely used in recent recommender systems, where negative sampling plays an important role. Existing negative sampling methods restrict the relationship between nodes as either hard positive pairs or hard negative pairs. This leads to the loss of structural information, and lacks the mechanism to generate positive pairs for nodes with few neighbors. To overcome limitations, we propose a novel soft link-based sampling method, namely MixDec Sampling, which consists of Mixup Sampling module and Decay Sampling module. The Mixup Sampling augments node features by synthesizing new nodes and soft links, which provides sufficient number of samples for nodes with few neighbors. The Decay Sampling strengthens the digestion of graph structure information by generating soft links for node embedding learning. To the best of our knowledge, we are the first to model sampling relationships between nodes by soft links in GNN-based recommender systems. Extensive experiments demonstrate that the proposed MixDec Sampling can significantly and consistently improve the recommendation performance of several representative GNN-based models on various recommendation benchmarks.
RECIST-Net: Lesion detection via grouping keypoints on RECIST-based annotation
Jul 19, 2021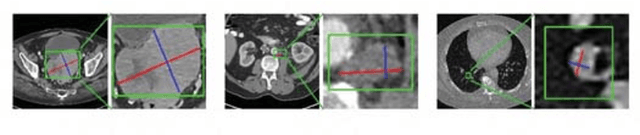
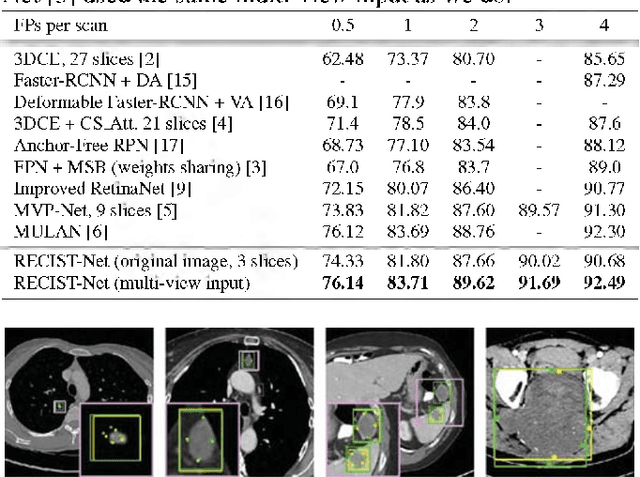
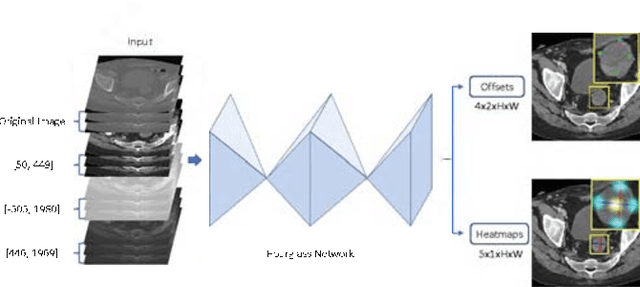
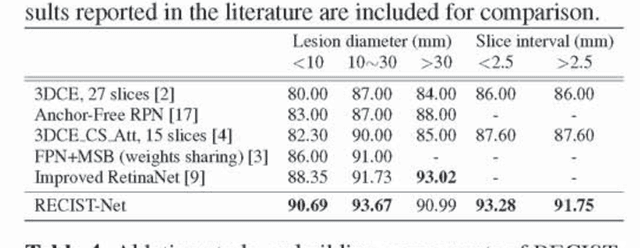
Abstract:Universal lesion detection in computed tomography (CT) images is an important yet challenging task due to the large variations in lesion type, size, shape, and appearance. Considering that data in clinical routine (such as the DeepLesion dataset) are usually annotated with a long and a short diameter according to the standard of Response Evaluation Criteria in Solid Tumors (RECIST) diameters, we propose RECIST-Net, a new approach to lesion detection in which the four extreme points and center point of the RECIST diameters are detected. By detecting a lesion as keypoints, we provide a more conceptually straightforward formulation for detection, and overcome several drawbacks (e.g., requiring extensive effort in designing data-appropriate anchors and losing shape information) of existing bounding-box-based methods while exploring a single-task, one-stage approach compared to other RECIST-based approaches. Experiments show that RECIST-Net achieves a sensitivity of 92.49% at four false positives per image, outperforming other recent methods including those using multi-task learning.
Online Disease Self-diagnosis with Inductive Heterogeneous Graph Convolutional Networks
Sep 06, 2020



Abstract:We propose a Healthcare Graph Convolutional Network (HealGCN) to offer disease self-diagnosis service for online users, based on the Electronic Healthcare Records (EHRs). Two main challenges are focused in this paper for online disease self-diagnosis: (1) serving cold-start users via graph convolutional networks and (2) handling scarce clinical description via a symptom retrieval system. To this end, we first organize the EHR data into a heterogeneous graph that is capable of modeling complex interactions among users, symptoms and diseases, and tailor the graph representation learning towards disease diagnosis with an inductive learning paradigm. Then, we build a disease self-diagnosis system with a corresponding EHR Graph-based Symptom Retrieval System (GraphRet) that can search and provide a list of relevant alternative symptoms by tracing the predefined meta-paths. GraphRet helps enrich the seed symptom set through the EHR graph, resulting in better reasoning ability of our HealGCN model, when confronting users with scarce descriptions. At last, we validate our model on a large-scale EHR dataset, the superior performance does confirm our model's effectiveness in practice.
Modeling e-Learners' Cognitive and Metacognitive Strategy in Comparative Question Solving
Jun 04, 2019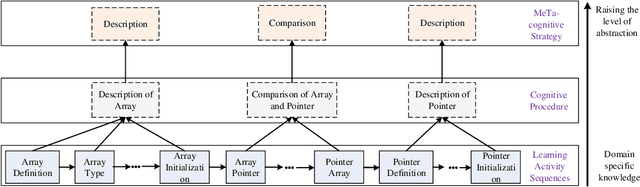
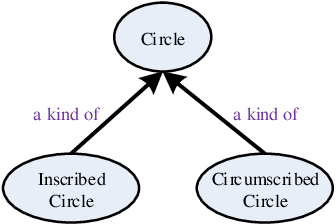
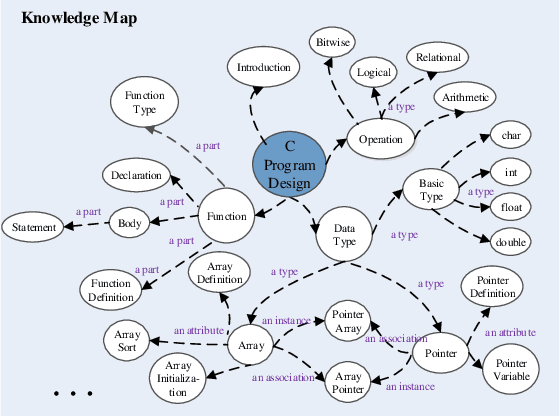
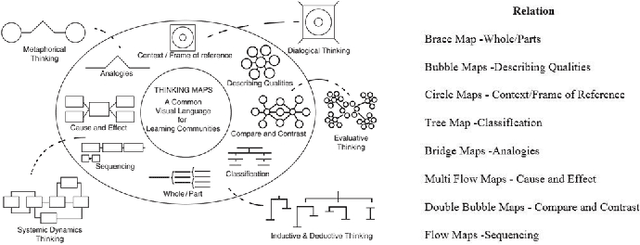
Abstract:Cognitive and metacognitive strategy had demonstrated a significant role in self-regulated learning (SRL), and an appropriate use of strategies is beneficial to effective learning or question-solving tasks during a human-computer interaction process. This paper proposes a novel method combining Knowledge Map (KM) based data mining technique with Thinking Map (TM) to detect learner's cognitive and metacognitive strategy in the question-solving scenario. In particular, a graph-based mining algorithm is designed to facilitate our proposed method, which can automatically map cognitive strategy to metacognitive strategy with raising abstraction level, and make the cognitive and metacognitive process viewable, which acts like a reverse engineering engine to explain how a learner thinks when solving a question. Additionally, we develop an online learning environment system for participants to learn and record their behaviors. To corroborate the effectiveness of our approach and algorithm, we conduct experiments recruiting 173 postgraduate and undergraduate students, and they were asked to complete a question-solving task, such as "What are similarities and differences between array and pointer?" from "The C Programming Language" course and "What are similarities and differences between packet switching and circuit switching?" from "Computer Network Principle" course. The mined strategies patterns results are encouraging and supported well our proposed method.
Measuring Patient Similarities via a Deep Architecture with Medical Concept Embedding
Feb 09, 2019



Abstract:Evaluating the clinical similarities between pairwise patients is a fundamental problem in healthcare informatics. A proper patient similarity measure enables various downstream applications, such as cohort study and treatment comparative effectiveness research. One major carrier for conducting patient similarity research is Electronic Health Records(EHRs), which are usually heterogeneous, longitudinal, and sparse. Though existing studies on learning patient similarity from EHRs have shown being useful in solving real clinical problems, their applicability is limited due to the lack of medical interpretations. Moreover, most previous methods assume a vector-based representation for patients, which typically requires aggregation of medical events over a certain time period. As a consequence, temporal information will be lost. In this paper, we propose a patient similarity evaluation framework based on the temporal matching of longitudinal patient EHRs. Two efficient methods are presented, unsupervised and supervised, both of which preserve the temporal properties in EHRs. The supervised scheme takes a convolutional neural network architecture and learns an optimal representation of patient clinical records with medical concept embedding. The empirical results on real-world clinical data demonstrate substantial improvement over the baselines. We make our code and sample data available for further study.
Safe Medicine Recommendation via Medical Knowledge Graph Embedding
Oct 26, 2017



Abstract:Most of the existing medicine recommendation systems that are mainly based on electronic medical records (EMRs) are significantly assisting doctors to make better clinical decisions benefiting both patients and caregivers. Even though the growth of EMRs is at a lighting fast speed in the era of big data, content limitations in EMRs restrain the existed recommendation systems to reflect relevant medical facts, such as drug-drug interactions. Many medical knowledge graphs that contain drug-related information, such as DrugBank, may give hope for the recommendation systems. However, the direct use of these knowledge graphs in the systems suffers from robustness caused by the incompleteness of the graphs. To address these challenges, we stand on recent advances in graph embedding learning techniques and propose a novel framework, called Safe Medicine Recommendation (SMR), in this paper. Specifically, SMR first constructs a high-quality heterogeneous graph by bridging EMRs (MIMIC-III) and medical knowledge graphs (ICD-9 ontology and DrugBank). Then, SMR jointly embeds diseases, medicines, patients, and their corresponding relations into a shared lower dimensional space. Finally, SMR uses the embeddings to decompose the medicine recommendation into a link prediction process while considering the patient's diagnoses and adverse drug reactions. To our best knowledge, SMR is the first to learn embeddings of a patient-disease-medicine graph for medicine recommendation in the world. Extensive experiments on real datasets are conducted to evaluate the effectiveness of proposed framework.
On Constrained Spectral Clustering and Its Applications
Sep 21, 2012



Abstract:Constrained clustering has been well-studied for algorithms such as $K$-means and hierarchical clustering. However, how to satisfy many constraints in these algorithmic settings has been shown to be intractable. One alternative to encode many constraints is to use spectral clustering, which remains a developing area. In this paper, we propose a flexible framework for constrained spectral clustering. In contrast to some previous efforts that implicitly encode Must-Link and Cannot-Link constraints by modifying the graph Laplacian or constraining the underlying eigenspace, we present a more natural and principled formulation, which explicitly encodes the constraints as part of a constrained optimization problem. Our method offers several practical advantages: it can encode the degree of belief in Must-Link and Cannot-Link constraints; it guarantees to lower-bound how well the given constraints are satisfied using a user-specified threshold; it can be solved deterministically in polynomial time through generalized eigendecomposition. Furthermore, by inheriting the objective function from spectral clustering and encoding the constraints explicitly, much of the existing analysis of unconstrained spectral clustering techniques remains valid for our formulation. We validate the effectiveness of our approach by empirical results on both artificial and real datasets. We also demonstrate an innovative use of encoding large number of constraints: transfer learning via constraints.
A Reconstruction Error Formulation for Semi-Supervised Multi-task and Multi-view Learning
Feb 04, 2012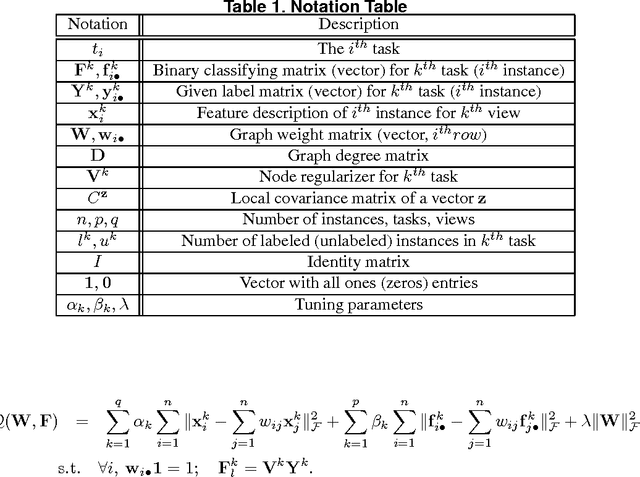
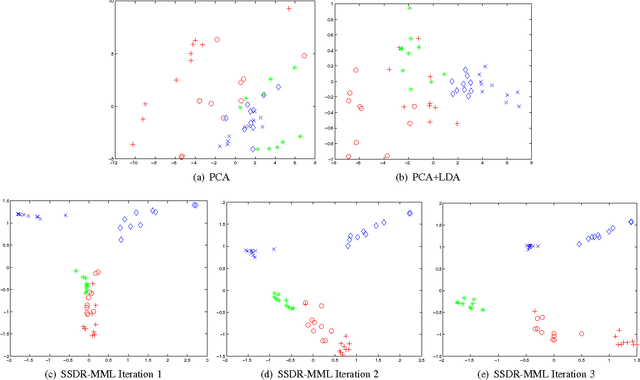
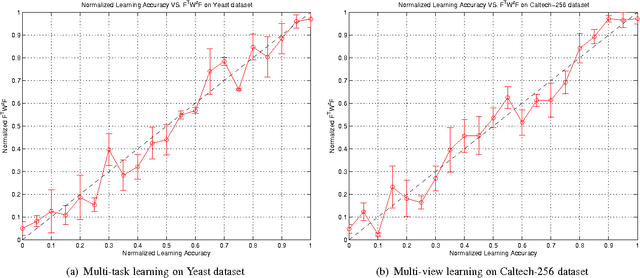
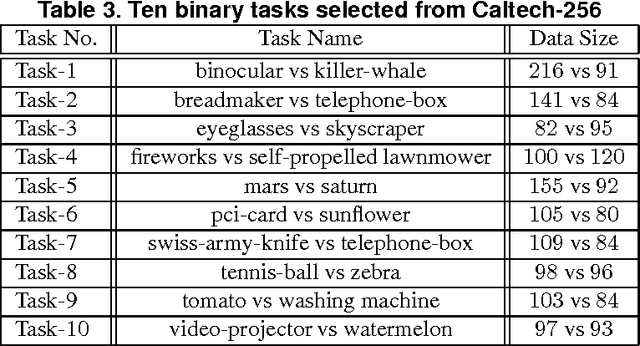
Abstract:A significant challenge to make learning techniques more suitable for general purpose use is to move beyond i) complete supervision, ii) low dimensional data, iii) a single task and single view per instance. Solving these challenges allows working with "Big Data" problems that are typically high dimensional with multiple (but possibly incomplete) labelings and views. While other work has addressed each of these problems separately, in this paper we show how to address them together, namely semi-supervised dimension reduction for multi-task and multi-view learning (SSDR-MML), which performs optimization for dimension reduction and label inference in semi-supervised setting. The proposed framework is designed to handle both multi-task and multi-view learning settings, and can be easily adapted to many useful applications. Information obtained from all tasks and views is combined via reconstruction errors in a linear fashion that can be efficiently solved using an alternating optimization scheme. Our formulation has a number of advantages. We explicitly model the information combining mechanism as a data structure (a weight/nearest-neighbor matrix) which allows investigating fundamental questions in multi-task and multi-view learning. We address one such question by presenting a general measure to quantify the success of simultaneous learning of multiple tasks or from multiple views. We show that our SSDR-MML approach can outperform many state-of-the-art baseline methods and demonstrate the effectiveness of connecting dimension reduction and learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge