Bizhe Bai
Learning to Explore with Parameter-Space Noise: A Deep Dive into Parameter-Space Noise for Reinforcement Learning with Verifiable Rewards
Jan 30, 2026Abstract:Reinforcement Learning with Verifiable Rewards (RLVR) improves LLM reasoning, yet growing evidence indicates an exploration ceiling: it often reweights existing solution traces rather than discovering new strategies, limiting gains under large sampling budgets (e.g., pass-at-256). We address this limitation with PSN-RLVR, which perturbs policy parameters before rollout generation to induce temporally consistent, trajectory-level exploration that better preserves long-horizon chain-of-thought coherence than action-space noise. To mitigate the resulting sampling-update mismatch, we incorporate truncated importance sampling (TIS). To avoid expensive KL-based adaptive noise control, we propose a computationally efficient real-time adaptive noise scheduler driven by a lightweight surrogate that combines semantic diversity with normalized self-certainty. Instantiated on GRPO, a widely used RLVR method, PSN-GRPO consistently expands the effective reasoning capability boundary across multiple mathematical reasoning benchmarks and model families, yielding higher pass-at-k under large sampling budgets and outperforming prior exploration-oriented RLVR methods (e.g., Pass-at-k-style training) while remaining orthogonal and thus composable for additional gains.
M-GRPO: Stabilizing Self-Supervised Reinforcement Learning for Large Language Models with Momentum-Anchored Policy Optimization
Dec 15, 2025Abstract:Self-supervised reinforcement learning (RL) presents a promising approach for enhancing the reasoning capabilities of Large Language Models (LLMs) without reliance on expensive human-annotated data. However, we find that existing methods suffer from a critical failure mode under long-horizon training: a "policy collapse" where performance precipitously degrades. We diagnose this instability and demonstrate that simply scaling the number of rollouts -- a common strategy to improve performance -- only delays, but does not prevent, this collapse. To counteract this instability, we first introduce M-GRPO (Momentum-Anchored Group Relative Policy Optimization), a framework that leverages a slowly evolving momentum model to provide a stable training target. In addition, we identify that this process is often accompanied by a rapid collapse in policy entropy, resulting in a prematurely confident and suboptimal policy. To specifically address this issue, we propose a second contribution: an adaptive filtering method based on the interquartile range (IQR) that dynamically prunes low-entropy trajectories, preserving essential policy diversity. Our extensive experiments on multiple reasoning benchmarks demonstrate that M-GRPO stabilizes the training process while the IQR filter prevents premature convergence. The combination of these two innovations leads to superior training stability and state-of-the-art performance.
Think Twice, Act Once: Token-Aware Compression and Action Reuse for Efficient Inference in Vision-Language-Action Models
May 27, 2025



Abstract:Vision-Language-Action (VLA) models have emerged as a powerful paradigm for general-purpose robot control through natural language instructions. However, their high inference cost-stemming from large-scale token computation and autoregressive decoding-poses significant challenges for real-time deployment and edge applications. While prior work has primarily focused on architectural optimization, we take a different perspective by identifying a dual form of redundancy in VLA models: (i) high similarity across consecutive action steps, and (ii) substantial redundancy in visual tokens. Motivated by these observations, we propose FlashVLA, the first training-free and plug-and-play acceleration framework that enables action reuse in VLA models. FlashVLA improves inference efficiency through a token-aware action reuse mechanism that avoids redundant decoding across stable action steps, and an information-guided visual token selection strategy that prunes low-contribution tokens. Extensive experiments on the LIBERO benchmark show that FlashVLA reduces FLOPs by 55.7% and latency by 36.0%, with only a 0.7% drop in task success rate. These results demonstrate the effectiveness of FlashVLA in enabling lightweight, low-latency VLA inference without retraining.
Local Information Matters: Inference Acceleration For Grounded Conversation Generation Models Through Adaptive Local-Aware Token Pruning
Apr 01, 2025
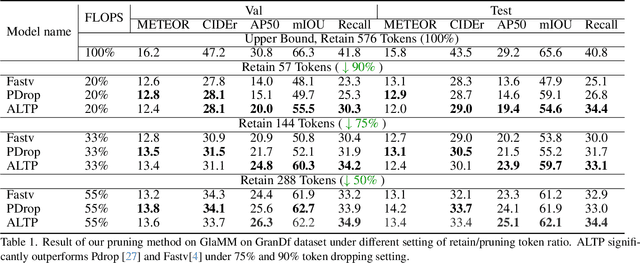


Abstract:Grounded Conversation Generation (GCG) is an emerging vision-language task that requires models to generate natural language responses seamlessly intertwined with corresponding object segmentation masks. Recent models, such as GLaMM and OMG-LLaVA, achieve pixel-level grounding but incur significant computational costs due to processing a large number of visual tokens. Existing token pruning methods, like FastV and PyramidDrop, fail to preserve the local visual features critical for accurate grounding, leading to substantial performance drops in GCG tasks. To address this, we propose Adaptive Local-Aware Token Pruning (ALTP), a simple yet effective framework that accelerates GCG models by prioritizing local object information. ALTP introduces two key components: (1) Detail Density Capture (DDC), which uses superpixel segmentation to retain tokens in object-centric regions, preserving fine-grained details, and (2) Dynamic Density Formation (DDF), which dynamically allocates tokens based on information density, ensuring higher retention in semantically rich areas. Extensive experiments on the GranDf dataset demonstrate that ALTP significantly outperforms existing token pruning methods, such as FastV and PyramidDrop, on both GLaMM and OMG-LLaVA models. Notably, when applied to GLaMM, ALTP achieves a 90% reduction in visual tokens with a 4.9% improvement in AP50 and a 5.0% improvement in Recall compared to PyramidDrop. Similarly, on OMG-LLaVA, ALTP improves AP by 2.1% and mIOU by 3.0% at a 90% token reduction compared with PDrop.
Cross-Phase Mutual Learning Framework for Pulmonary Embolism Identification on Non-Contrast CT Scans
Jul 16, 2024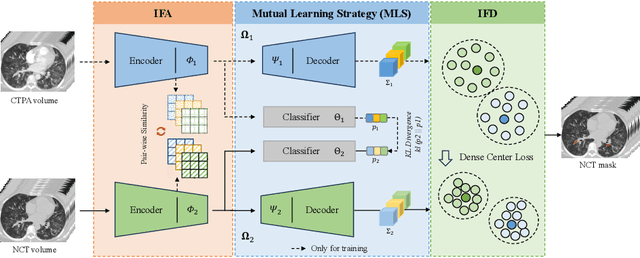
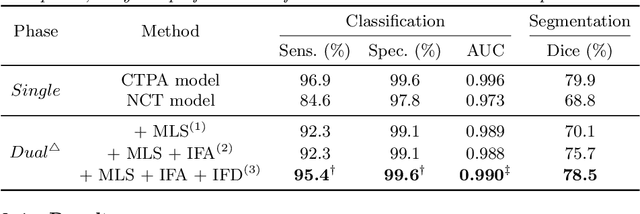
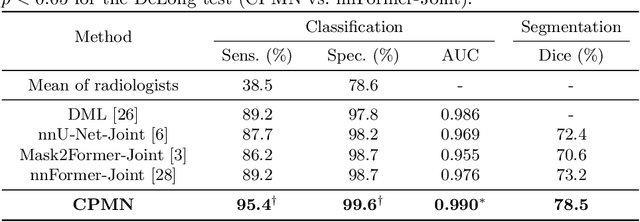
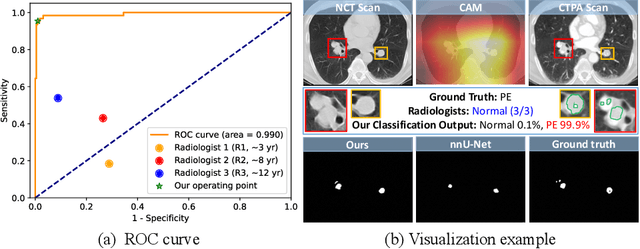
Abstract:Pulmonary embolism (PE) is a life-threatening condition where rapid and accurate diagnosis is imperative yet difficult due to predominantly atypical symptomatology. Computed tomography pulmonary angiography (CTPA) is acknowledged as the gold standard imaging tool in clinics, yet it can be contraindicated for emergency department (ED) patients and represents an onerous procedure, thus necessitating PE identification through non-contrast CT (NCT) scans. In this work, we explore the feasibility of applying a deep-learning approach to NCT scans for PE identification. We propose a novel Cross-Phase Mutual learNing framework (CPMN) that fosters knowledge transfer from CTPA to NCT, while concurrently conducting embolism segmentation and abnormality classification in a multi-task manner. The proposed CPMN leverages the Inter-Feature Alignment (IFA) strategy that enhances spatial contiguity and mutual learning between the dual-pathway network, while the Intra-Feature Discrepancy (IFD) strategy can facilitate precise segmentation of PE against complex backgrounds for single-pathway networks. For a comprehensive assessment of the proposed approach, a large-scale dual-phase dataset containing 334 PE patients and 1,105 normal subjects has been established. Experimental results demonstrate that CPMN achieves the leading identification performance, which is 95.4\% and 99.6\% in patient-level sensitivity and specificity on NCT scans, indicating the potential of our approach as an economical, accessible, and precise tool for PE identification in clinical practice.
Pseudo Label-Guided Data Fusion and Output Consistency for Semi-Supervised Medical Image Segmentation
Nov 17, 2023



Abstract:Supervised learning algorithms based on Convolutional Neural Networks have become the benchmark for medical image segmentation tasks, but their effectiveness heavily relies on a large amount of labeled data. However, annotating medical image datasets is a laborious and time-consuming process. Inspired by semi-supervised algorithms that use both labeled and unlabeled data for training, we propose the PLGDF framework, which builds upon the mean teacher network for segmenting medical images with less annotation. We propose a novel pseudo-label utilization scheme, which combines labeled and unlabeled data to augment the dataset effectively. Additionally, we enforce the consistency between different scales in the decoder module of the segmentation network and propose a loss function suitable for evaluating the consistency. Moreover, we incorporate a sharpening operation on the predicted results, further enhancing the accuracy of the segmentation. Extensive experiments on three publicly available datasets demonstrate that the PLGDF framework can largely improve performance by incorporating the unlabeled data. Meanwhile, our framework yields superior performance compared to six state-of-the-art semi-supervised learning methods. The codes of this study are available at https://github.com/ortonwang/PLGDF.
The Multi-modality Cell Segmentation Challenge: Towards Universal Solutions
Aug 10, 2023Abstract:Cell segmentation is a critical step for quantitative single-cell analysis in microscopy images. Existing cell segmentation methods are often tailored to specific modalities or require manual interventions to specify hyperparameters in different experimental settings. Here, we present a multi-modality cell segmentation benchmark, comprising over 1500 labeled images derived from more than 50 diverse biological experiments. The top participants developed a Transformer-based deep-learning algorithm that not only exceeds existing methods, but can also be applied to diverse microscopy images across imaging platforms and tissue types without manual parameter adjustments. This benchmark and the improved algorithm offer promising avenues for more accurate and versatile cell analysis in microscopy imaging.
SurgT challenge: Benchmark of Soft-Tissue Trackers for Robotic Surgery
Feb 28, 2023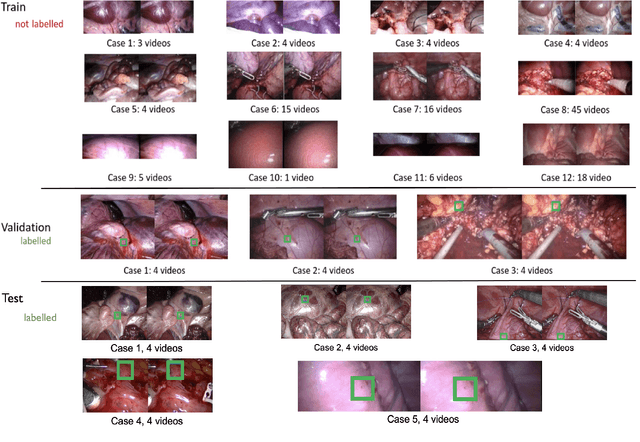
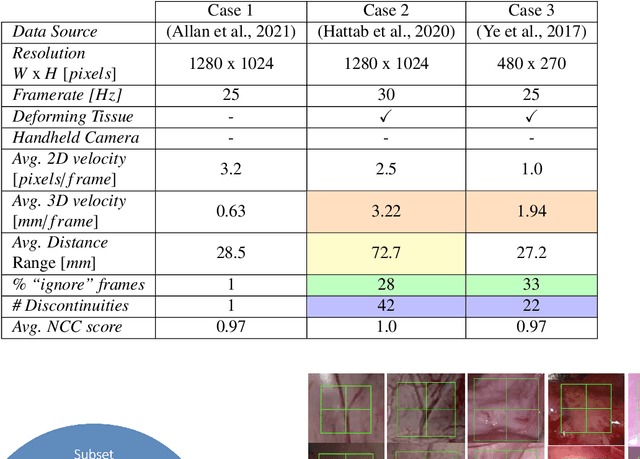
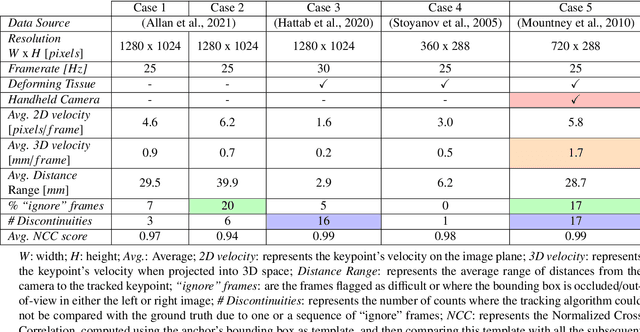
Abstract:This paper introduces the "SurgT: Surgical Tracking" challenge which was organised in conjunction with the 25th International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI 2022). There were two purposes for the creation of this challenge: (1) the establishment of the first standardised benchmark for the research community to assess soft-tissue trackers; and (2) to encourage the development of unsupervised deep learning methods, given the lack of annotated data in surgery. A dataset of 157 stereo endoscopic videos from 20 clinical cases, along with stereo camera calibration parameters, have been provided. The participants were tasked with the development of algorithms to track a bounding box on stereo endoscopic videos. At the end of the challenge, the developed methods were assessed on a previously hidden test subset. This assessment uses benchmarking metrics that were purposely developed for this challenge and are now available online. The teams were ranked according to their Expected Average Overlap (EAO) score, which is a weighted average of the Intersection over Union (IoU) scores. The performance evaluation study verifies the efficacy of unsupervised deep learning algorithms in tracking soft-tissue. The best-performing method achieved an EAO score of 0.583 in the test subset. The dataset and benchmarking tool created for this challenge have been made publicly available. This challenge is expected to contribute to the development of autonomous robotic surgery and other digital surgical technologies.
FetReg2021: A Challenge on Placental Vessel Segmentation and Registration in Fetoscopy
Jun 30, 2022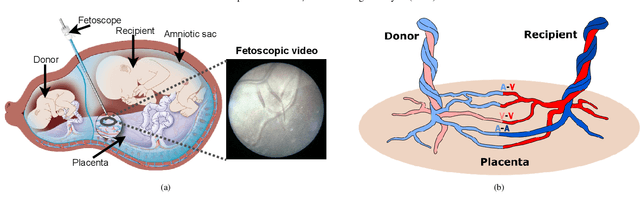
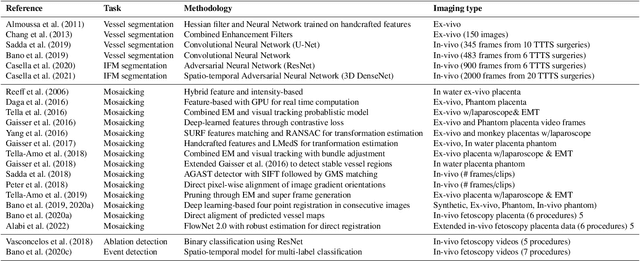
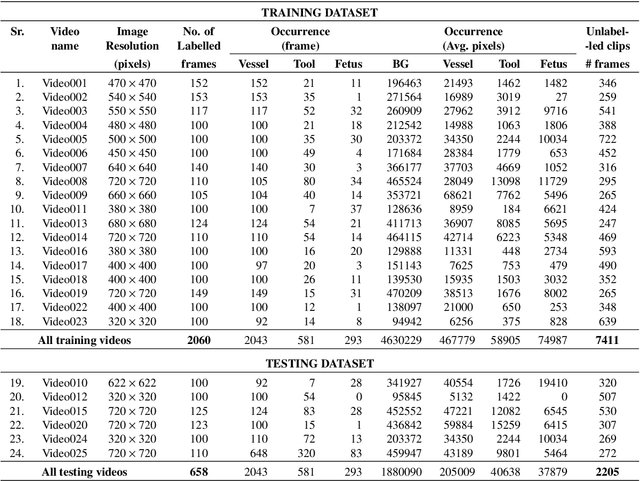
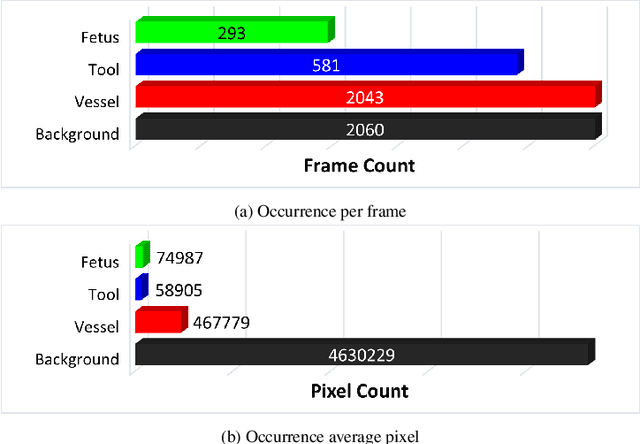
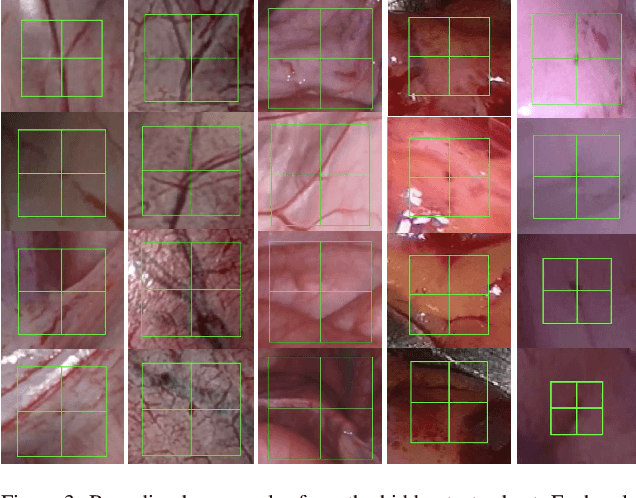
Abstract:Fetoscopy laser photocoagulation is a widely adopted procedure for treating Twin-to-Twin Transfusion Syndrome (TTTS). The procedure involves photocoagulation pathological anastomoses to regulate blood exchange among twins. The procedure is particularly challenging due to the limited field of view, poor manoeuvrability of the fetoscope, poor visibility, and variability in illumination. These challenges may lead to increased surgery time and incomplete ablation. Computer-assisted intervention (CAI) can provide surgeons with decision support and context awareness by identifying key structures in the scene and expanding the fetoscopic field of view through video mosaicking. Research in this domain has been hampered by the lack of high-quality data to design, develop and test CAI algorithms. Through the Fetoscopic Placental Vessel Segmentation and Registration (FetReg2021) challenge, which was organized as part of the MICCAI2021 Endoscopic Vision challenge, we released the first largescale multicentre TTTS dataset for the development of generalized and robust semantic segmentation and video mosaicking algorithms. For this challenge, we released a dataset of 2060 images, pixel-annotated for vessels, tool, fetus and background classes, from 18 in-vivo TTTS fetoscopy procedures and 18 short video clips. Seven teams participated in this challenge and their model performance was assessed on an unseen test dataset of 658 pixel-annotated images from 6 fetoscopic procedures and 6 short clips. The challenge provided an opportunity for creating generalized solutions for fetoscopic scene understanding and mosaicking. In this paper, we present the findings of the FetReg2021 challenge alongside reporting a detailed literature review for CAI in TTTS fetoscopy. Through this challenge, its analysis and the release of multi-centre fetoscopic data, we provide a benchmark for future research in this field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge