Infant Brain Mri Segmentation
Papers and Code
Deep learning-based neurodevelopmental assessment in preterm infants
Jan 17, 2026Preterm infants (born between 28 and 37 weeks of gestation) face elevated risks of neurodevelopmental delays, making early identification crucial for timely intervention. While deep learning-based volumetric segmentation of brain MRI scans offers a promising avenue for assessing neonatal neurodevelopment, achieving accurate segmentation of white matter (WM) and gray matter (GM) in preterm infants remains challenging due to their comparable signal intensities (isointense appearance) on MRI during early brain development. To address this, we propose a novel segmentation neural network, named Hierarchical Dense Attention Network. Our architecture incorporates a 3D spatial-channel attention mechanism combined with an attention-guided dense upsampling strategy to enhance feature discrimination in low-contrast volumetric data. Quantitative experiments demonstrate that our method achieves superior segmentation performance compared to state-of-the-art baselines, effectively tackling the challenge of isointense tissue differentiation. Furthermore, application of our algorithm confirms that WM and GM volumes in preterm infants are significantly lower than those in term infants, providing additional imaging evidence of the neurodevelopmental delays associated with preterm birth. The code is available at: https://github.com/ICL-SUST/HDAN.
Comparison of different segmentation algorithms on brain volume and fractal dimension in infant brain MRIs
Dec 13, 2025Accurate segmentation of infant brain MRI is essential for quantifying developmental changes in structure and complexity. However, ongoing myelination and reduced tissue contrast make automated segmentation particularly challenging. This study systematically compared segmentation accuracy and its impact on volumetric and fractal dimension (FD) estimates in infant brain MRI using the Baby Open Brains (BOB) dataset (71 scans, 1-9 months). Two methods, SynthSeg and SamSeg, were evaluated against expert annotations using Dice, Intersection over Union, 95th-percentile Hausdorff distance, and Normalised Mutual Information. SynthSeg outperformed SamSeg across all quality metrics (mean Dice > 0.8 for major regions) and provided volumetric estimates closely matching the manual reference (mean +4% [-28% - 71%]). SamSeg systematically overestimated ventricular and whole-brain volumes (mean +76% [-12% - 190%]). Segmentation accuracy improved with age, consistent with increasing tissue contrast during myelination. Fractal dimension a(FD) nalyses revealed significant regional differences between SynthSeg and expert segmentations, and Bland-Altman limits of agreement indicated that segmentation-related FD variability exceeded most group differences reported in developmental cohorts. Volume and FD deviations were positively correlated across structures, indicating that segmentation bias directly affects FD estimation. Overall, SynthSeg provided the most reliable volumetric and FD results for paediatric MRI, yet small morphological differences in volume and FD should be interpreted with caution due to segmentation-related uncertainty.
Bilateral Hippocampi Segmentation in Low Field MRIs Using Mutual Feature Learning via Dual-Views
Oct 23, 2024



Accurate hippocampus segmentation in brain MRI is critical for studying cognitive and memory functions and diagnosing neurodevelopmental disorders. While high-field MRIs provide detailed imaging, low-field MRIs are more accessible and cost-effective, which eliminates the need for sedation in children, though they often suffer from lower image quality. In this paper, we present a novel deep-learning approach for the automatic segmentation of bilateral hippocampi in low-field MRIs. Extending recent advancements in infant brain segmentation to underserved communities through the use of low-field MRIs ensures broader access to essential diagnostic tools, thereby supporting better healthcare outcomes for all children. Inspired by our previous work, Co-BioNet, the proposed model employs a dual-view structure to enable mutual feature learning via high-frequency masking, enhancing segmentation accuracy by leveraging complementary information from different perspectives. Extensive experiments demonstrate that our method provides reliable segmentation outcomes for hippocampal analysis in low-resource settings. The code is publicly available at: https://github.com/himashi92/LoFiHippSeg.
Cas-DiffCom: Cascaded diffusion model for infant longitudinal super-resolution 3D medical image completion
Feb 21, 2024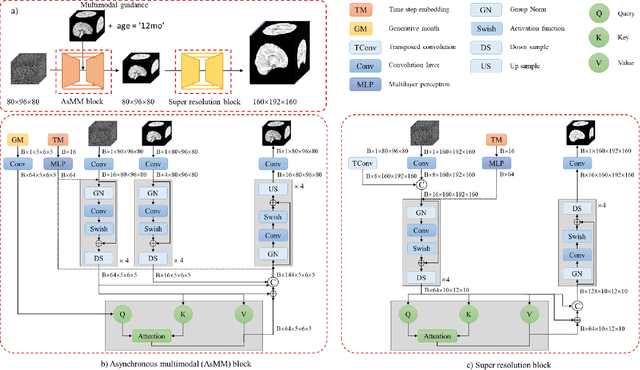
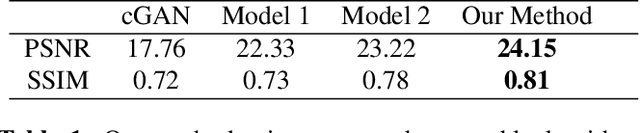
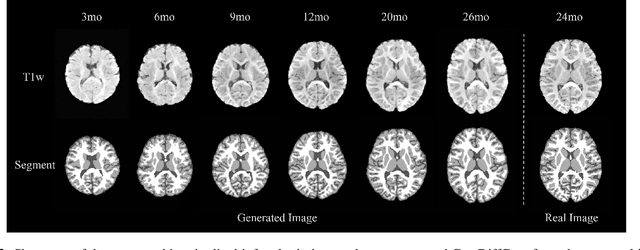
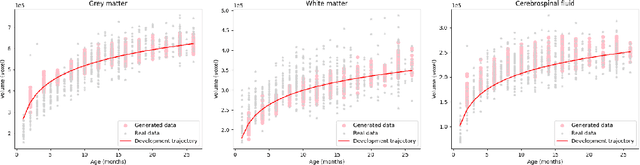
Early infancy is a rapid and dynamic neurodevelopmental period for behavior and neurocognition. Longitudinal magnetic resonance imaging (MRI) is an effective tool to investigate such a crucial stage by capturing the developmental trajectories of the brain structures. However, longitudinal MRI acquisition always meets a serious data-missing problem due to participant dropout and failed scans, making longitudinal infant brain atlas construction and developmental trajectory delineation quite challenging. Thanks to the development of an AI-based generative model, neuroimage completion has become a powerful technique to retain as much available data as possible. However, current image completion methods usually suffer from inconsistency within each individual subject in the time dimension, compromising the overall quality. To solve this problem, our paper proposed a two-stage cascaded diffusion model, Cas-DiffCom, for dense and longitudinal 3D infant brain MRI completion and super-resolution. We applied our proposed method to the Baby Connectome Project (BCP) dataset. The experiment results validate that Cas-DiffCom achieves both individual consistency and high fidelity in longitudinal infant brain image completion. We further applied the generated infant brain images to two downstream tasks, brain tissue segmentation and developmental trajectory delineation, to declare its task-oriented potential in the neuroscience field.
3D Masked Autoencoding and Pseudo-labeling for Domain Adaptive Segmentation of Heterogeneous Infant Brain MRI
Mar 16, 2023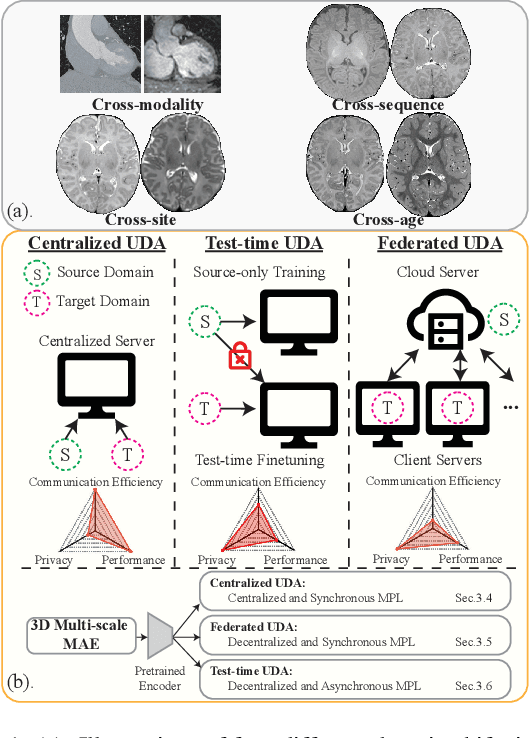
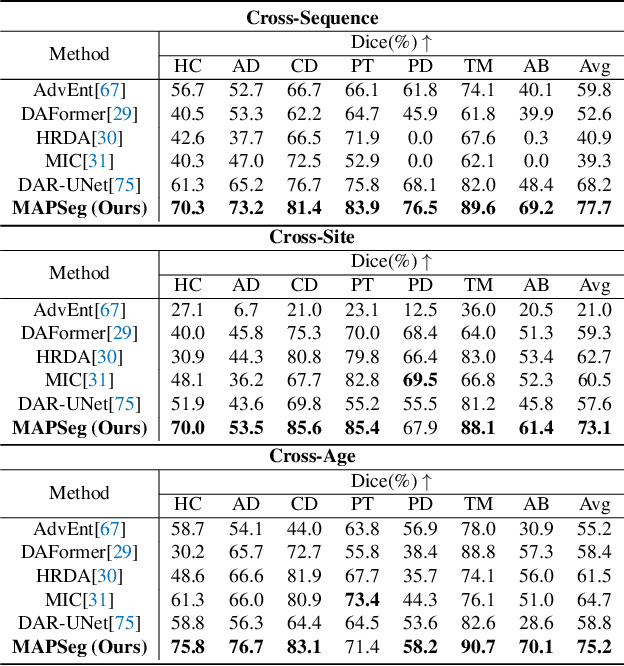
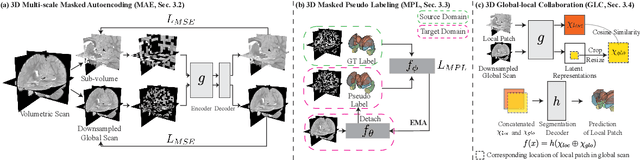

Robust segmentation of infant brain MRI across multiple ages, modalities, and sites remains challenging due to the intrinsic heterogeneity caused by different MRI scanners, vendors, or acquisition sequences, as well as varying stages of neurodevelopment. To address this challenge, previous studies have explored domain adaptation (DA) algorithms from various perspectives, including feature alignment, entropy minimization, contrast synthesis (style transfer), and pseudo-labeling. This paper introduces a novel framework called MAPSeg (Masked Autoencoding and Pseudo-labelling Segmentation) to address the challenges of cross-age, cross-modality, and cross-site segmentation of subcortical regions in infant brain MRI. Utilizing 3D masked autoencoding as well as masked pseudo-labeling, the model is able to jointly learn from labeled source domain data and unlabeled target domain data. We evaluated our framework on expert-annotated datasets acquired from different ages and sites. MAPSeg consistently outperformed other methods, including previous state-of-the-art supervised baselines, domain generalization, and domain adaptation frameworks in segmenting subcortical regions regardless of age, modality, or acquisition site. The code and pretrained encoder will be publicly available at https://github.com/XuzheZ/MAPSeg
Two Independent Teachers are Better Role Model
Jun 09, 2023Recent deep learning models have attracted substantial attention in infant brain analysis. These models have performed state-of-the-art performance, such as semi-supervised techniques (e.g., Temporal Ensembling, mean teacher). However, these models depend on an encoder-decoder structure with stacked local operators to gather long-range information, and the local operators limit the efficiency and effectiveness. Besides, the $MRI$ data contain different tissue properties ($TPs$) such as $T1$ and $T2$. One major limitation of these models is that they use both data as inputs to the segment process, i.e., the models are trained on the dataset once, and it requires much computational and memory requirements during inference. In this work, we address the above limitations by designing a new deep-learning model, called 3D-DenseUNet, which works as adaptable global aggregation blocks in down-sampling to solve the issue of spatial information loss. The self-attention module connects the down-sampling blocks to up-sampling blocks, and integrates the feature maps in three dimensions of spatial and channel, effectively improving the representation potential and discriminating ability of the model. Additionally, we propose a new method called Two Independent Teachers ($2IT$), that summarizes the model weights instead of label predictions. Each teacher model is trained on different types of brain data, $T1$ and $T2$, respectively. Then, a fuse model is added to improve test accuracy and enable training with fewer parameters and labels compared to the Temporal Ensembling method without modifying the network architecture. Empirical results demonstrate the effectiveness of the proposed method.
DAM-AL: Dilated Attention Mechanism with Attention Loss for 3D Infant Brain Image Segmentation
Dec 27, 2021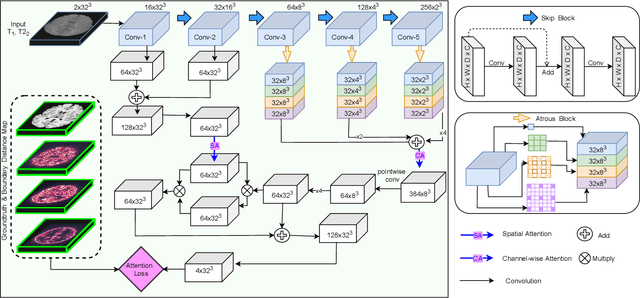


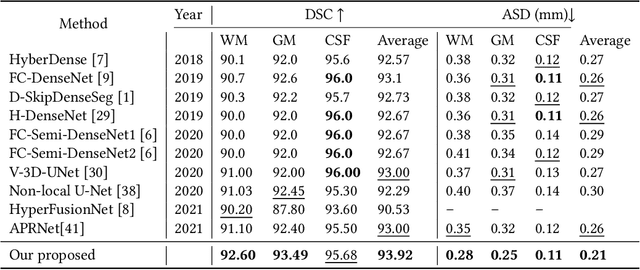
While Magnetic Resonance Imaging (MRI) has played an essential role in infant brain analysis, segmenting MRI into a number of tissues such as gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) is crucial and complex due to the extremely low intensity contrast between tissues at around 6-9 months of age as well as amplified noise, myelination, and incomplete volume. In this paper, we tackle those limitations by developing a new deep learning model, named DAM-AL, which contains two main contributions, i.e., dilated attention mechanism and hard-case attention loss. Our DAM-AL network is designed with skip block layers and atrous block convolution. It contains both channel-wise attention at high-level context features and spatial attention at low-level spatial structural features. Our attention loss consists of two terms corresponding to region information and hard samples attention. Our proposed DAM-AL has been evaluated on the infant brain iSeg 2017 dataset and the experiments have been conducted on both validation and testing sets. We have benchmarked DAM-AL on Dice coefficient and ASD metrics and compared it with state-of-the-art methods.
Local Spatiotemporal Representation Learning for Longitudinally-consistent Neuroimage Analysis
Jun 09, 2022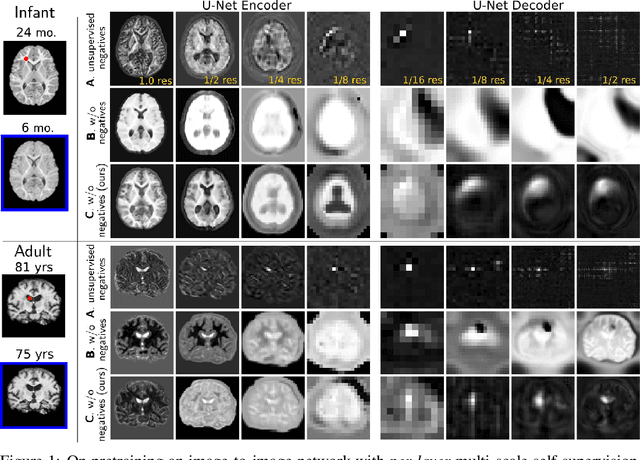
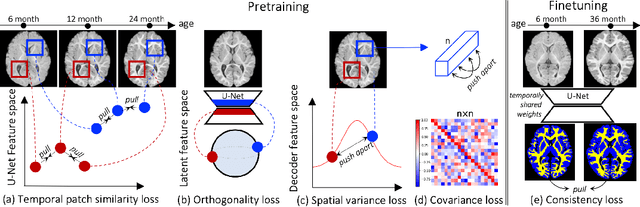

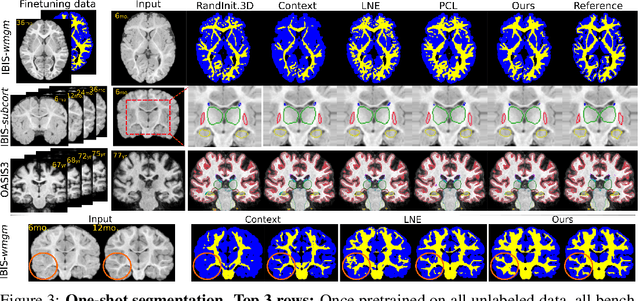
Recent self-supervised advances in medical computer vision exploit global and local anatomical self-similarity for pretraining prior to downstream tasks such as segmentation. However, current methods assume i.i.d. image acquisition, which is invalid in clinical study designs where follow-up longitudinal scans track subject-specific temporal changes. Further, existing self-supervised methods for medically-relevant image-to-image architectures exploit only spatial or temporal self-similarity and only do so via a loss applied at a single image-scale, with naive multi-scale spatiotemporal extensions collapsing to degenerate solutions. To these ends, this paper makes two contributions: (1) It presents a local and multi-scale spatiotemporal representation learning method for image-to-image architectures trained on longitudinal images. It exploits the spatiotemporal self-similarity of learned multi-scale intra-subject features for pretraining and develops several feature-wise regularizations that avoid collapsed identity representations; (2) During finetuning, it proposes a surprisingly simple self-supervised segmentation consistency regularization to exploit intra-subject correlation. Benchmarked in the one-shot segmentation setting, the proposed framework outperforms both well-tuned randomly-initialized baselines and current self-supervised techniques designed for both i.i.d. and longitudinal datasets. These improvements are demonstrated across both longitudinal neurodegenerative adult MRI and developing infant brain MRI and yield both higher performance and longitudinal consistency.
Infant brain MRI segmentation with dilated convolution pyramid downsampling and self-attention
Dec 29, 2019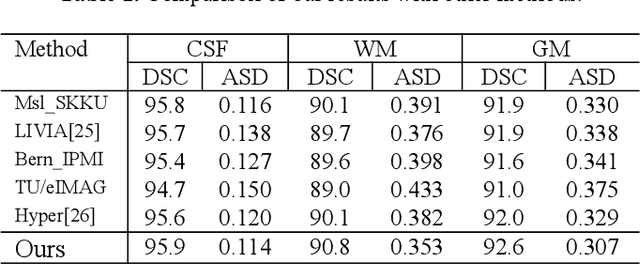
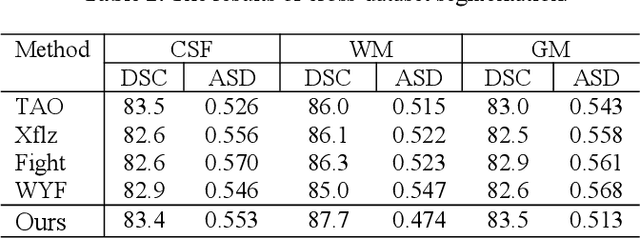
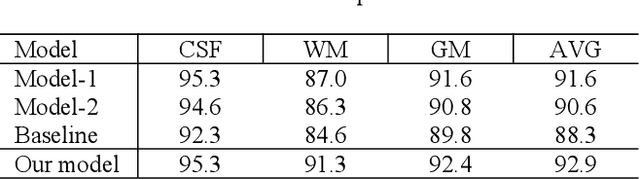
In this paper, we propose a dual aggregation network to adaptively aggregate different information in infant brain MRI segmentation. More precisely, we added two modules based on 3D-UNet to better model information at different levels and locations. The dilated convolution pyramid downsampling module is mainly to solve the problem of loss of spatial information on the downsampling process, and it can effectively save details while reducing the resolution. The self-attention module can integrate the remote dependence on the feature maps in two dimensions of spatial and channel, effectively improving the representation ability and discriminating ability of the model. Our results are compared to the winners of iseg2017's first evaluation, the DICE ratio of WM and GM increased by 0.7%, and CSF is comparable.In the latest evaluation of the iseg-2019 cross-dataset challenge,we achieve the first place in the DICE of WM and GM, and the DICE of CSF is second.
InfiNet: Fully Convolutional Networks for Infant Brain MRI Segmentation
Oct 11, 2018



We present a novel, parameter-efficient and practical fully convolutional neural network architecture, termed InfiNet, aimed at voxel-wise semantic segmentation of infant brain MRI images at iso-intense stage, which can be easily extended for other segmentation tasks involving multi-modalities. InfiNet consists of double encoder arms for T1 and T2 input scans that feed into a joint-decoder arm that terminates in the classification layer. The novelty of InfiNet lies in the manner in which the decoder upsamples lower resolution input feature map(s) from multiple encoder arms. Specifically, the pooled indices computed in the max-pooling layers of each of the encoder blocks are related to the corresponding decoder block to perform non-linear learning-free upsampling. The sparse maps are concatenated with intermediate encoder representations (skip connections) and convolved with trainable filters to produce dense feature maps. InfiNet is trained end-to-end to optimize for the Generalized Dice Loss, which is well-suited for high class imbalance. InfiNet achieves the whole-volume segmentation in under 50 seconds and we demonstrate competitive performance against multiple state-of-the art deep architectures and their multi-modal variants.
* 4 pages, 3 figures, conference, IEEE ISBI, 2018
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge