Dinh-Hieu Hoang
NeIn: Telling What You Don't Want
Sep 09, 2024



Abstract:Negation is a fundamental linguistic concept used by humans to convey information that they do not desire. Despite this, there has been minimal research specifically focused on negation within vision-language tasks. This lack of research means that vision-language models (VLMs) may struggle to understand negation, implying that they struggle to provide accurate results. One barrier to achieving human-level intelligence is the lack of a standard collection by which research into negation can be evaluated. This paper presents the first large-scale dataset, Negative Instruction (NeIn), for studying negation within the vision-language domain. Our dataset comprises 530,694 quadruples, i.e., source image, original caption, negative sentence, and target image in total, including 495,694 queries for training and 35,000 queries for benchmarking across multiple vision-language tasks. Specifically, we automatically generate NeIn based on a large, existing vision-language dataset, MS-COCO, via two steps: generation and filtering. During the generation phase, we leverage two VLMs, BLIP and MagicBrush, to generate the target image and a negative clause that expresses the content of the source image. In the subsequent filtering phase, we apply BLIP to remove erroneous samples. Additionally, we introduce an evaluation protocol for negation understanding of image editing models. Extensive experiments using our dataset across multiple VLMs for instruction-based image editing tasks demonstrate that even recent state-of-the-art VLMs struggle to understand negative queries. The project page is: https://tanbuinhat.github.io/NeIn/
TSRNet: Simple Framework for Real-time ECG Anomaly Detection with Multimodal Time and Spectrogram Restoration Network
Dec 15, 2023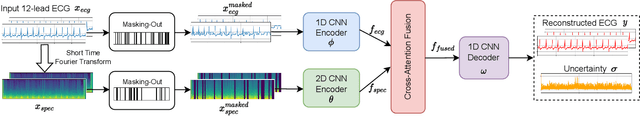
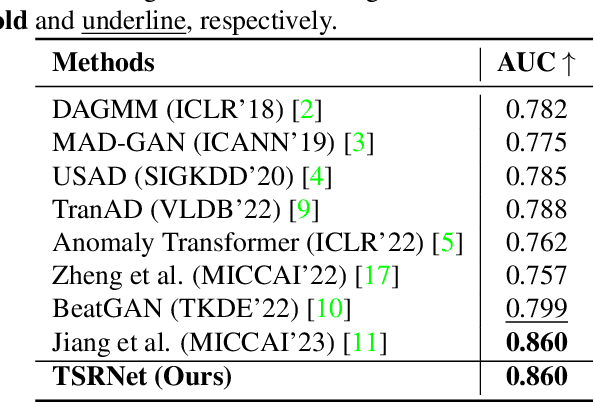
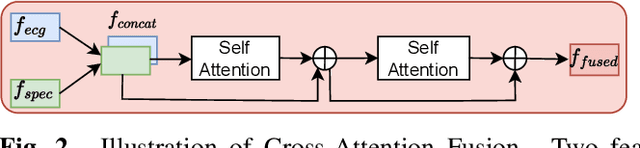
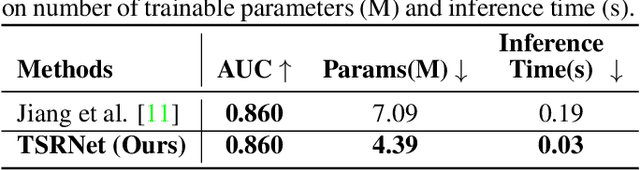
Abstract:The electrocardiogram (ECG) is a valuable signal used to assess various aspects of heart health, such as heart rate and rhythm. It plays a crucial role in identifying cardiac conditions and detecting anomalies in ECG data. However, distinguishing between normal and abnormal ECG signals can be a challenging task. In this paper, we propose an approach that leverages anomaly detection to identify unhealthy conditions using solely normal ECG data for training. Furthermore, to enhance the information available and build a robust system, we suggest considering both the time series and time-frequency domain aspects of the ECG signal. As a result, we introduce a specialized network called the Multimodal Time and Spectrogram Restoration Network (TSRNet) designed specifically for detecting anomalies in ECG signals. TSRNet falls into the category of restoration-based anomaly detection and draws inspiration from both the time series and spectrogram domains. By extracting representations from both domains, TSRNet effectively captures the comprehensive characteristics of the ECG signal. This approach enables the network to learn robust representations with superior discrimination abilities, allowing it to distinguish between normal and abnormal ECG patterns more effectively. Furthermore, we introduce a novel inference method, termed Peak-based Error, that specifically focuses on ECG peaks, a critical component in detecting abnormalities. The experimental result on the large-scale dataset PTB-XL has demonstrated the effectiveness of our approach in ECG anomaly detection, while also prioritizing efficiency by minimizing the number of trainable parameters. Our code is available at https://github.com/UARK-AICV/TSRNet.
SAM3D: Segment Anything Model in Volumetric Medical Images
Sep 07, 2023



Abstract:Image segmentation is a critical task in medical image analysis, providing valuable information that helps to make an accurate diagnosis. In recent years, deep learning-based automatic image segmentation methods have achieved outstanding results in medical images. In this paper, inspired by the Segment Anything Model (SAM), a foundation model that has received much attention for its impressive accuracy and powerful generalization ability in 2D still image segmentation, we propose a SAM3D that targets at 3D volumetric medical images and utilizes the pre-trained features from the SAM encoder to capture meaningful representations of input images. Different from other existing SAM-based volumetric segmentation methods that perform the segmentation by dividing the volume into a set of 2D slices, our model takes the whole 3D volume image as input and processes it simply and effectively that avoids training a significant number of parameters. Extensive experiments are conducted on multiple medical image datasets to demonstrate that our network attains competitive results compared with other state-of-the-art methods in 3D medical segmentation tasks while being significantly efficient in terms of parameters.
MEGANet: Multi-Scale Edge-Guided Attention Network for Weak Boundary Polyp Segmentation
Sep 06, 2023



Abstract:Efficient polyp segmentation in healthcare plays a critical role in enabling early diagnosis of colorectal cancer. However, the segmentation of polyps presents numerous challenges, including the intricate distribution of backgrounds, variations in polyp sizes and shapes, and indistinct boundaries. Defining the boundary between the foreground (i.e. polyp itself) and the background (surrounding tissue) is difficult. To mitigate these challenges, we propose Multi-Scale Edge-Guided Attention Network (MEGANet) tailored specifically for polyp segmentation within colonoscopy images. This network draws inspiration from the fusion of a classical edge detection technique with an attention mechanism. By combining these techniques, MEGANet effectively preserves high-frequency information, notably edges and boundaries, which tend to erode as neural networks deepen. MEGANet is designed as an end-to-end framework, encompassing three key modules: an encoder, which is responsible for capturing and abstracting the features from the input image, a decoder, which focuses on salient features, and the Edge-Guided Attention module (EGA) that employs the Laplacian Operator to accentuate polyp boundaries. Extensive experiments, both qualitative and quantitative, on five benchmark datasets, demonstrate that our EGANet outperforms other existing SOTA methods under six evaluation metrics. Our code is available at \url{https://github.com/DinhHieuHoang/MEGANet}
DAM-AL: Dilated Attention Mechanism with Attention Loss for 3D Infant Brain Image Segmentation
Dec 27, 2021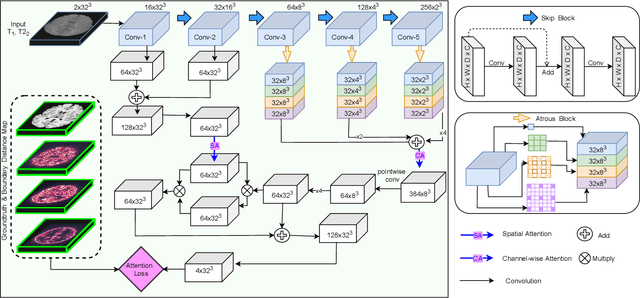


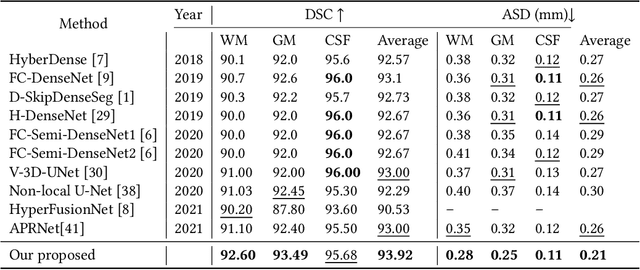
Abstract:While Magnetic Resonance Imaging (MRI) has played an essential role in infant brain analysis, segmenting MRI into a number of tissues such as gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) is crucial and complex due to the extremely low intensity contrast between tissues at around 6-9 months of age as well as amplified noise, myelination, and incomplete volume. In this paper, we tackle those limitations by developing a new deep learning model, named DAM-AL, which contains two main contributions, i.e., dilated attention mechanism and hard-case attention loss. Our DAM-AL network is designed with skip block layers and atrous block convolution. It contains both channel-wise attention at high-level context features and spatial attention at low-level spatial structural features. Our attention loss consists of two terms corresponding to region information and hard samples attention. Our proposed DAM-AL has been evaluated on the infant brain iSeg 2017 dataset and the experiments have been conducted on both validation and testing sets. We have benchmarked DAM-AL on Dice coefficient and ASD metrics and compared it with state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge