Gia-Han Diep
Point-Unet: A Context-aware Point-based Neural Network for Volumetric Segmentation
Mar 16, 2022
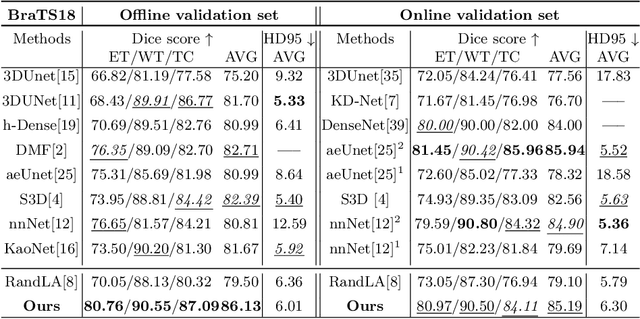

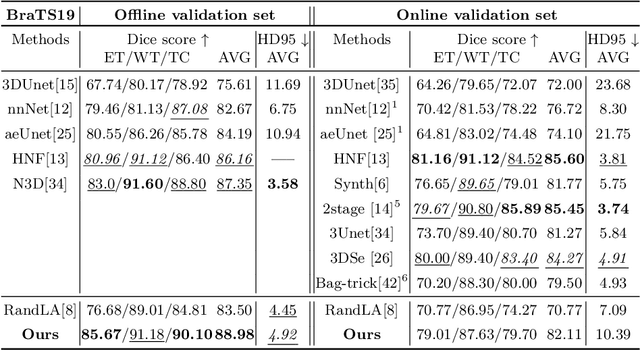
Abstract:Medical image analysis using deep learning has recently been prevalent, showing great performance for various downstream tasks including medical image segmentation and its sibling, volumetric image segmentation. Particularly, a typical volumetric segmentation network strongly relies on a voxel grid representation which treats volumetric data as a stack of individual voxel `slices', which allows learning to segment a voxel grid to be as straightforward as extending existing image-based segmentation networks to the 3D domain. However, using a voxel grid representation requires a large memory footprint, expensive test-time and limiting the scalability of the solutions. In this paper, we propose Point-Unet, a novel method that incorporates the efficiency of deep learning with 3D point clouds into volumetric segmentation. Our key idea is to first predict the regions of interest in the volume by learning an attentional probability map, which is then used for sampling the volume into a sparse point cloud that is subsequently segmented using a point-based neural network. We have conducted the experiments on the medical volumetric segmentation task with both a small-scale dataset Pancreas and large-scale datasets BraTS18, BraTS19, and BraTS20 challenges. A comprehensive benchmark on different metrics has shown that our context-aware Point-Unet robustly outperforms the SOTA voxel-based networks at both accuracies, memory usage during training, and time consumption during testing. Our code is available at https://github.com/VinAIResearch/Point-Unet.
DAM-AL: Dilated Attention Mechanism with Attention Loss for 3D Infant Brain Image Segmentation
Dec 27, 2021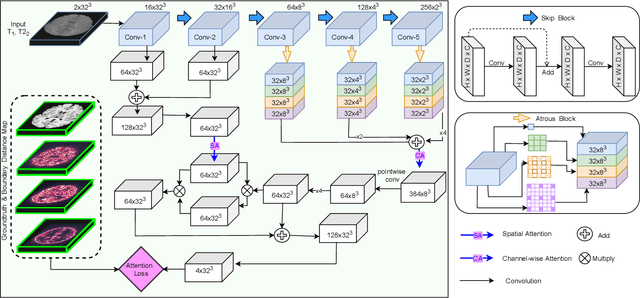


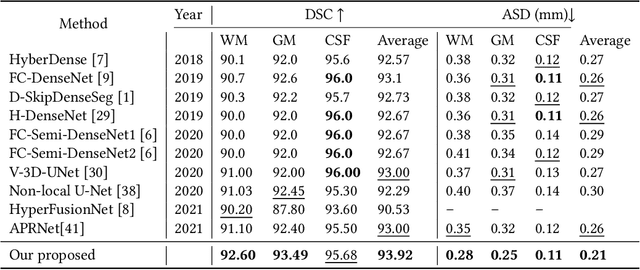
Abstract:While Magnetic Resonance Imaging (MRI) has played an essential role in infant brain analysis, segmenting MRI into a number of tissues such as gray matter (GM), white matter (WM), and cerebrospinal fluid (CSF) is crucial and complex due to the extremely low intensity contrast between tissues at around 6-9 months of age as well as amplified noise, myelination, and incomplete volume. In this paper, we tackle those limitations by developing a new deep learning model, named DAM-AL, which contains two main contributions, i.e., dilated attention mechanism and hard-case attention loss. Our DAM-AL network is designed with skip block layers and atrous block convolution. It contains both channel-wise attention at high-level context features and spatial attention at low-level spatial structural features. Our attention loss consists of two terms corresponding to region information and hard samples attention. Our proposed DAM-AL has been evaluated on the infant brain iSeg 2017 dataset and the experiments have been conducted on both validation and testing sets. We have benchmarked DAM-AL on Dice coefficient and ASD metrics and compared it with state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge