Ngoc-Vuong Ho
FG-CXR: A Radiologist-Aligned Gaze Dataset for Enhancing Interpretability in Chest X-Ray Report Generation
Nov 23, 2024



Abstract:Developing an interpretable system for generating reports in chest X-ray (CXR) analysis is becoming increasingly crucial in Computer-aided Diagnosis (CAD) systems, enabling radiologists to comprehend the decisions made by these systems. Despite the growth of diverse datasets and methods focusing on report generation, there remains a notable gap in how closely these models' generated reports align with the interpretations of real radiologists. In this study, we tackle this challenge by initially introducing Fine-Grained CXR (FG-CXR) dataset, which provides fine-grained paired information between the captions generated by radiologists and the corresponding gaze attention heatmaps for each anatomy. Unlike existing datasets that include a raw sequence of gaze alongside a report, with significant misalignment between gaze location and report content, our FG-CXR dataset offers a more grained alignment between gaze attention and diagnosis transcript. Furthermore, our analysis reveals that simply applying black-box image captioning methods to generate reports cannot adequately explain which information in CXR is utilized and how long needs to attend to accurately generate reports. Consequently, we propose a novel explainable radiologist's attention generator network (Gen-XAI) that mimics the diagnosis process of radiologists, explicitly constraining its output to closely align with both radiologist's gaze attention and transcript. Finally, we perform extensive experiments to illustrate the effectiveness of our method. Our datasets and checkpoint is available at https://github.com/UARK-AICV/FG-CXR.
Point-Unet: A Context-aware Point-based Neural Network for Volumetric Segmentation
Mar 16, 2022
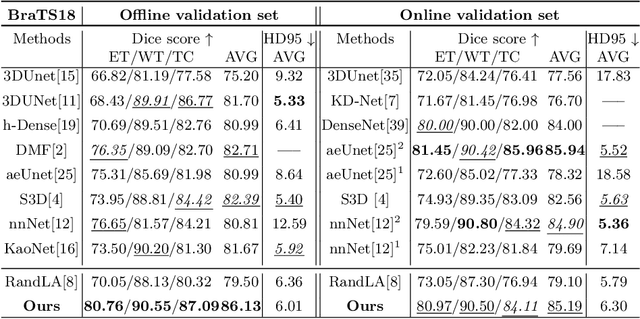

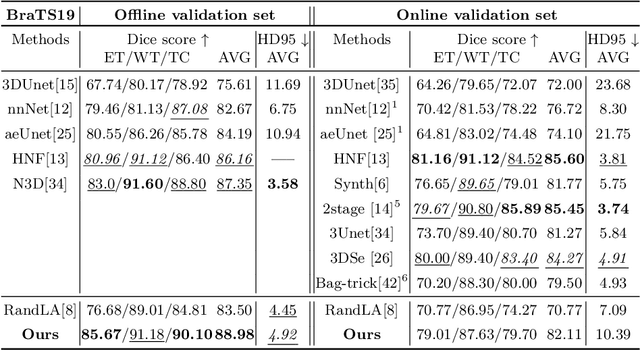
Abstract:Medical image analysis using deep learning has recently been prevalent, showing great performance for various downstream tasks including medical image segmentation and its sibling, volumetric image segmentation. Particularly, a typical volumetric segmentation network strongly relies on a voxel grid representation which treats volumetric data as a stack of individual voxel `slices', which allows learning to segment a voxel grid to be as straightforward as extending existing image-based segmentation networks to the 3D domain. However, using a voxel grid representation requires a large memory footprint, expensive test-time and limiting the scalability of the solutions. In this paper, we propose Point-Unet, a novel method that incorporates the efficiency of deep learning with 3D point clouds into volumetric segmentation. Our key idea is to first predict the regions of interest in the volume by learning an attentional probability map, which is then used for sampling the volume into a sparse point cloud that is subsequently segmented using a point-based neural network. We have conducted the experiments on the medical volumetric segmentation task with both a small-scale dataset Pancreas and large-scale datasets BraTS18, BraTS19, and BraTS20 challenges. A comprehensive benchmark on different metrics has shown that our context-aware Point-Unet robustly outperforms the SOTA voxel-based networks at both accuracies, memory usage during training, and time consumption during testing. Our code is available at https://github.com/VinAIResearch/Point-Unet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge