Patel Brijesh
FG-CXR: A Radiologist-Aligned Gaze Dataset for Enhancing Interpretability in Chest X-Ray Report Generation
Nov 23, 2024



Abstract:Developing an interpretable system for generating reports in chest X-ray (CXR) analysis is becoming increasingly crucial in Computer-aided Diagnosis (CAD) systems, enabling radiologists to comprehend the decisions made by these systems. Despite the growth of diverse datasets and methods focusing on report generation, there remains a notable gap in how closely these models' generated reports align with the interpretations of real radiologists. In this study, we tackle this challenge by initially introducing Fine-Grained CXR (FG-CXR) dataset, which provides fine-grained paired information between the captions generated by radiologists and the corresponding gaze attention heatmaps for each anatomy. Unlike existing datasets that include a raw sequence of gaze alongside a report, with significant misalignment between gaze location and report content, our FG-CXR dataset offers a more grained alignment between gaze attention and diagnosis transcript. Furthermore, our analysis reveals that simply applying black-box image captioning methods to generate reports cannot adequately explain which information in CXR is utilized and how long needs to attend to accurately generate reports. Consequently, we propose a novel explainable radiologist's attention generator network (Gen-XAI) that mimics the diagnosis process of radiologists, explicitly constraining its output to closely align with both radiologist's gaze attention and transcript. Finally, we perform extensive experiments to illustrate the effectiveness of our method. Our datasets and checkpoint is available at https://github.com/UARK-AICV/FG-CXR.
Multimodality Multi-Lead ECG Arrhythmia Classification using Self-Supervised Learning
Sep 30, 2022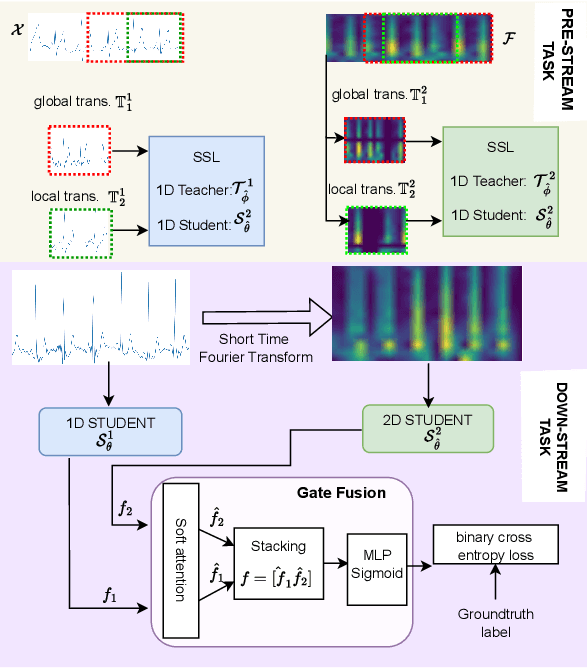
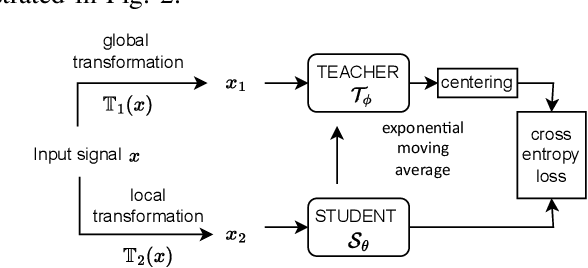
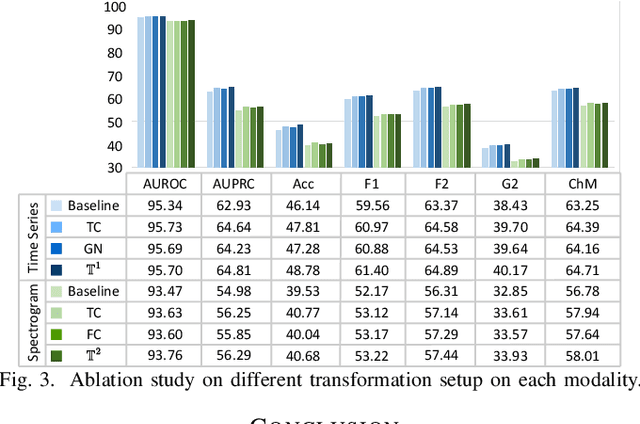
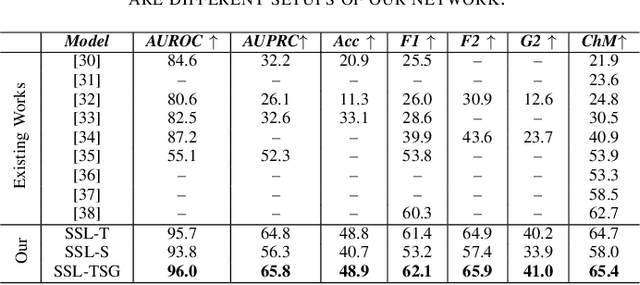
Abstract:Electrocardiogram (ECG) signal is one of the most effective sources of information mainly employed for the diagnosis and prediction of cardiovascular diseases (CVDs) connected with the abnormalities in heart rhythm. Clearly, single modality ECG (i.e. time series) cannot convey its complete characteristics, thus, exploiting both time and time-frequency modalities in the form of time-series data and spectrogram is needed. Leveraging the cutting-edge self-supervised learning (SSL) technique on unlabeled data, we propose SSL-based multimodality ECG classification. Our proposed network follows SSL learning paradigm and consists of two modules corresponding to pre-stream task, and down-stream task, respectively. In the SSL-pre-stream task, we utilize self-knowledge distillation (KD) techniques with no labeled data, on various transformations and in both time and frequency domains. In the down-stream task, which is trained on labeled data, we propose a gate fusion mechanism to fuse information from multimodality.To evaluate the effectiveness of our approach, ten-fold cross validation on the 12-lead PhysioNet 2020 dataset has been conducted.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge