Zhaolin Chen
Segment Any Tumour: An Uncertainty-Aware Vision Foundation Model for Whole-Body Analysis
Nov 11, 2025Abstract:Prompt-driven vision foundation models, such as the Segment Anything Model, have recently demonstrated remarkable adaptability in computer vision. However, their direct application to medical imaging remains challenging due to heterogeneous tissue structures, imaging artefacts, and low-contrast boundaries, particularly in tumours and cancer primaries leading to suboptimal segmentation in ambiguous or overlapping lesion regions. Here, we present Segment Any Tumour 3D (SAT3D), a lightweight volumetric foundation model designed to enable robust and generalisable tumour segmentation across diverse medical imaging modalities. SAT3D integrates a shifted-window vision transformer for hierarchical volumetric representation with an uncertainty-aware training pipeline that explicitly incorporates uncertainty estimates as prompts to guide reliable boundary prediction in low-contrast regions. Adversarial learning further enhances model performance for the ambiguous pathological regions. We benchmark SAT3D against three recent vision foundation models and nnUNet across 11 publicly available datasets, encompassing 3,884 tumour and cancer cases for training and 694 cases for in-distribution evaluation. Trained on 17,075 3D volume-mask pairs across multiple modalities and cancer primaries, SAT3D demonstrates strong generalisation and robustness. To facilitate practical use and clinical translation, we developed a 3D Slicer plugin that enables interactive, prompt-driven segmentation and visualisation using the trained SAT3D model. Extensive experiments highlight its effectiveness in improving segmentation accuracy under challenging and out-of-distribution scenarios, underscoring its potential as a scalable foundation model for medical image analysis.
Style Content Decomposition-based Data Augmentation for Domain Generalizable Medical Image Segmentation
Feb 28, 2025Abstract:Due to the domain shifts between training and testing medical images, learned segmentation models often experience significant performance degradation during deployment. In this paper, we first decompose an image into its style code and content map and reveal that domain shifts in medical images involve: \textbf{style shifts} (\emph{i.e.}, differences in image appearance) and \textbf{content shifts} (\emph{i.e.}, variations in anatomical structures), the latter of which has been largely overlooked. To this end, we propose \textbf{StyCona}, a \textbf{sty}le \textbf{con}tent decomposition-based data \textbf{a}ugmentation method that innovatively augments both image style and content within the rank-one space, for domain generalizable medical image segmentation. StyCona is a simple yet effective plug-and-play module that substantially improves model generalization without requiring additional training parameters or modifications to the segmentation model architecture. Experiments on cross-sequence, cross-center, and cross-modality medical image segmentation settings with increasingly severe domain shifts, demonstrate the effectiveness of StyCona and its superiority over state-of-the-arts. The code is available at https://github.com/Senyh/StyCona.
CodeBrain: Impute Any Brain MRI via Instance-specific Scalar-quantized Codes
Jan 30, 2025Abstract:MRI imputation aims to synthesize the missing modality from one or more available ones, which is highly desirable since it reduces scanning costs and delivers comprehensive MRI information to enhance clinical diagnosis. In this paper, we propose a unified model, CodeBrain, designed to adapt to various brain MRI imputation scenarios. The core design lies in casting various inter-modality transformations as a full-modality code prediction task. To this end, CodeBrain is trained in two stages: Reconstruction and Code Prediction. First, in the Reconstruction stage, we reconstruct each MRI modality, which is mapped into a shared latent space followed by a scalar quantization. Since such quantization is lossy and the code is low dimensional, another MRI modality belonging to the same subject is randomly selected to generate common features to supplement the code and boost the target reconstruction. In the second stage, we train another encoder by a customized grading loss to predict the full-modality codes from randomly masked MRI samples, supervised by the corresponding quantized codes generated from the first stage. In this way, the inter-modality transformation is achieved by mapping the instance-specific codes in a finite scalar space. We evaluated the proposed CodeBrain model on two public brain MRI datasets (i.e., IXI and BraTS 2023). Extensive experiments demonstrate that our CodeBrain model achieves superior imputation performance compared to four existing methods, establishing a new state of the art for unified brain MRI imputation. Codes will be released.
A Physics-based Generative Model to Synthesize Training Datasets for MRI-based Fat Quantification
Dec 11, 2024Abstract:Deep learning-based techniques have potential to optimize scan and post-processing times required for MRI-based fat quantification, but they are constrained by the lack of large training datasets. Generative models are a promising tool to perform data augmentation by synthesizing realistic datasets. However no previous methods have been specifically designed to generate datasets for quantitative MRI (q-MRI) tasks, where reference quantitative maps and large variability in scanning protocols are usually required. We propose a Physics-Informed Latent Diffusion Model (PI-LDM) to synthesize quantitative parameter maps jointly with customizable MR images by incorporating the signal generation model. We assessed the quality of PI-LDM's synthesized data using metrics such as the Fr\'echet Inception Distance (FID), obtaining comparable scores to state-of-the-art generative methods (FID: 0.0459). We also trained a U-Net for the MRI-based fat quantification task incorporating synthetic datasets. When we used a few real (10 subjects, $~200$ slices) and numerous synthetic samples ($>3000$), fat fraction at specific liver ROIs showed a low bias on data obtained using the same protocol than training data ($0.10\%$ at $\hbox{ROI}_1$, $0.12\%$ at $\hbox{ROI}_2$) and on data acquired with an alternative protocol ($0.14\%$ at $\hbox{ROI}_1$, $0.62\%$ at $\hbox{ROI}_2$). Future work will be to extend PI-LDM to other q-MRI applications.
Bilateral Hippocampi Segmentation in Low Field MRIs Using Mutual Feature Learning via Dual-Views
Oct 23, 2024



Abstract:Accurate hippocampus segmentation in brain MRI is critical for studying cognitive and memory functions and diagnosing neurodevelopmental disorders. While high-field MRIs provide detailed imaging, low-field MRIs are more accessible and cost-effective, which eliminates the need for sedation in children, though they often suffer from lower image quality. In this paper, we present a novel deep-learning approach for the automatic segmentation of bilateral hippocampi in low-field MRIs. Extending recent advancements in infant brain segmentation to underserved communities through the use of low-field MRIs ensures broader access to essential diagnostic tools, thereby supporting better healthcare outcomes for all children. Inspired by our previous work, Co-BioNet, the proposed model employs a dual-view structure to enable mutual feature learning via high-frequency masking, enhancing segmentation accuracy by leveraging complementary information from different perspectives. Extensive experiments demonstrate that our method provides reliable segmentation outcomes for hippocampal analysis in low-resource settings. The code is publicly available at: https://github.com/himashi92/LoFiHippSeg.
Deep kernel representations of latent space features for low-dose PET-MR imaging robust to variable dose reduction
Sep 10, 2024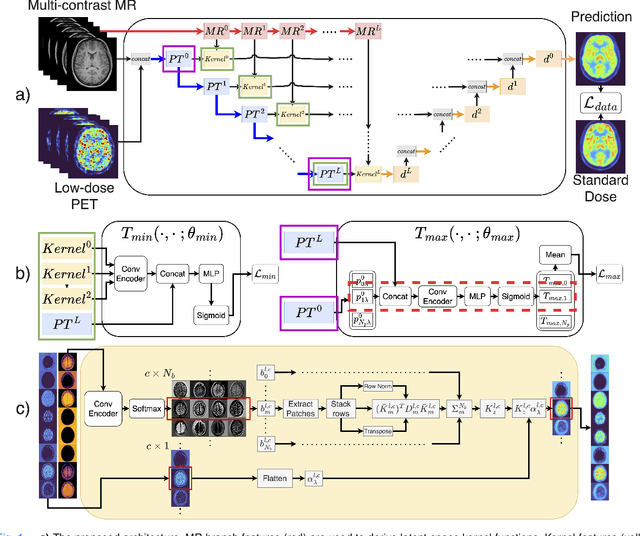
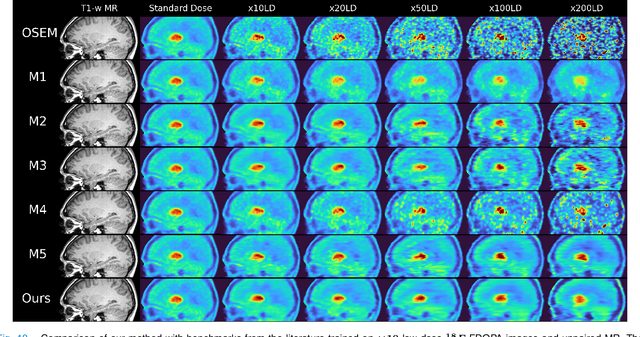
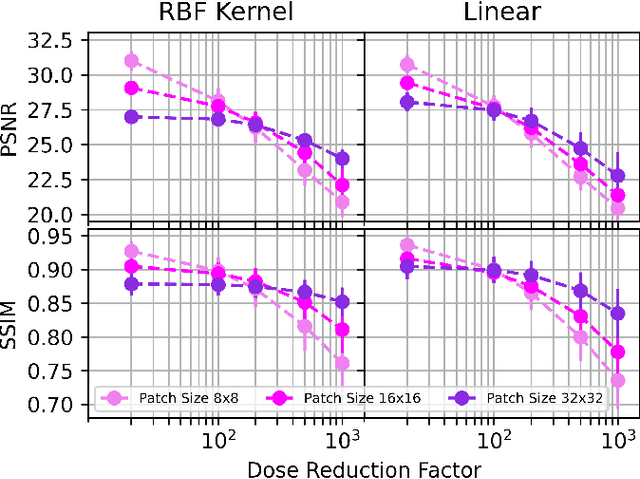
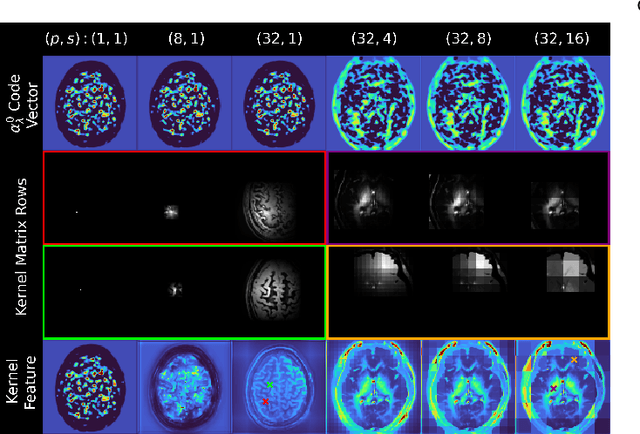
Abstract:Low-dose positron emission tomography (PET) image reconstruction methods have potential to significantly improve PET as an imaging modality. Deep learning provides a promising means of incorporating prior information into the image reconstruction problem to produce quantitatively accurate images from compromised signal. Deep learning-based methods for low-dose PET are generally poorly conditioned and perform unreliably on images with features not present in the training distribution. We present a method which explicitly models deep latent space features using a robust kernel representation, providing robust performance on previously unseen dose reduction factors. Additional constraints on the information content of deep latent features allow for tuning in-distribution accuracy and generalisability. Tests with out-of-distribution dose reduction factors ranging from $\times 10$ to $\times 1000$ and with both paired and unpaired MR, demonstrate significantly improved performance relative to conventional deep-learning methods trained using the same data. Code:https://github.com/cameronPain
SeCo-INR: Semantically Conditioned Implicit Neural Representations for Improved Medical Image Super-Resolution
Sep 02, 2024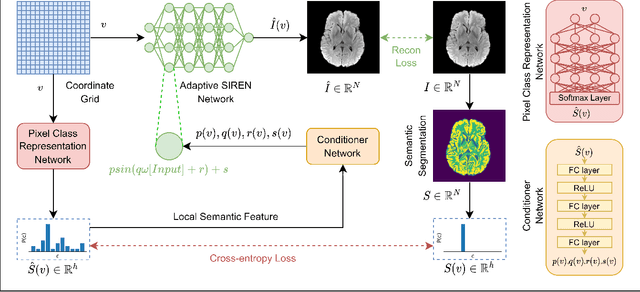
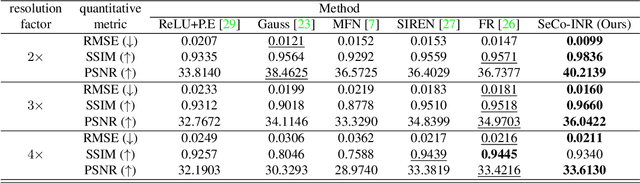
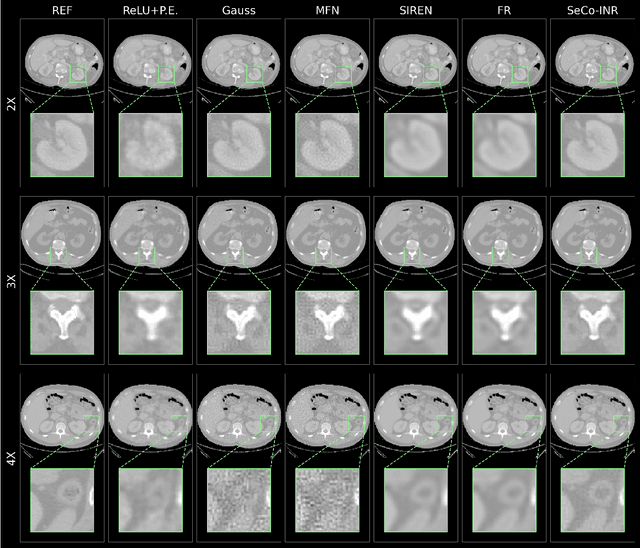
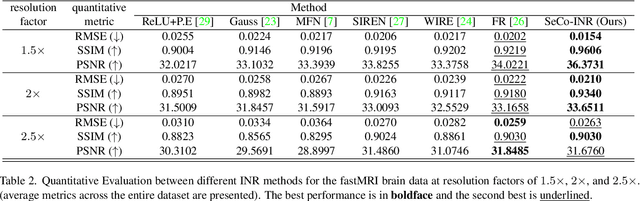
Abstract:Implicit Neural Representations (INRs) have recently advanced the field of deep learning due to their ability to learn continuous representations of signals without the need for large training datasets. Although INR methods have been studied for medical image super-resolution, their adaptability to localized priors in medical images has not been extensively explored. Medical images contain rich anatomical divisions that could provide valuable local prior information to enhance the accuracy and robustness of INRs. In this work, we propose a novel framework, referred to as the Semantically Conditioned INR (SeCo-INR), that conditions an INR using local priors from a medical image, enabling accurate model fitting and interpolation capabilities to achieve super-resolution. Our framework learns a continuous representation of the semantic segmentation features of a medical image and utilizes it to derive the optimal INR for each semantic region of the image. We tested our framework using several medical imaging modalities and achieved higher quantitative scores and more realistic super-resolution outputs compared to state-of-the-art methods.
McCaD: Multi-Contrast MRI Conditioned, Adaptive Adversarial Diffusion Model for High-Fidelity MRI Synthesis
Sep 01, 2024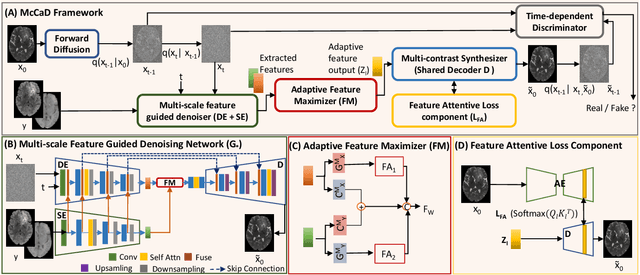



Abstract:Magnetic Resonance Imaging (MRI) is instrumental in clinical diagnosis, offering diverse contrasts that provide comprehensive diagnostic information. However, acquiring multiple MRI contrasts is often constrained by high costs, long scanning durations, and patient discomfort. Current synthesis methods, typically focused on single-image contrasts, fall short in capturing the collective nuances across various contrasts. Moreover, existing methods for multi-contrast MRI synthesis often fail to accurately map feature-level information across multiple imaging contrasts. We introduce McCaD (Multi-Contrast MRI Conditioned Adaptive Adversarial Diffusion), a novel framework leveraging an adversarial diffusion model conditioned on multiple contrasts for high-fidelity MRI synthesis. McCaD significantly enhances synthesis accuracy by employing a multi-scale, feature-guided mechanism, incorporating denoising and semantic encoders. An adaptive feature maximization strategy and a spatial feature-attentive loss have been introduced to capture more intrinsic features across multiple contrasts. This facilitates a precise and comprehensive feature-guided denoising process. Extensive experiments on tumor and healthy multi-contrast MRI datasets demonstrated that the McCaD outperforms state-of-the-art baselines quantitively and qualitatively. The code is provided with supplementary materials.
SAM-Med3D-MoE: Towards a Non-Forgetting Segment Anything Model via Mixture of Experts for 3D Medical Image Segmentation
Jul 06, 2024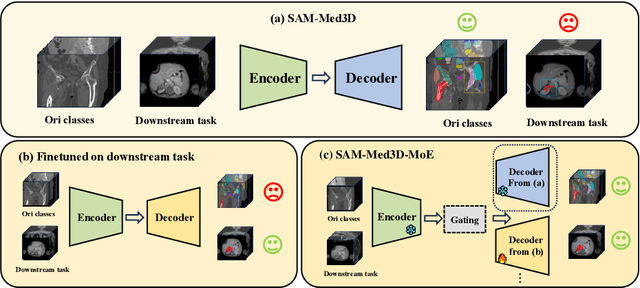

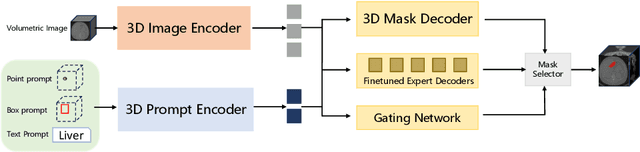

Abstract:Volumetric medical image segmentation is pivotal in enhancing disease diagnosis, treatment planning, and advancing medical research. While existing volumetric foundation models for medical image segmentation, such as SAM-Med3D and SegVol, have shown remarkable performance on general organs and tumors, their ability to segment certain categories in clinical downstream tasks remains limited. Supervised Finetuning (SFT) serves as an effective way to adapt such foundation models for task-specific downstream tasks but at the cost of degrading the general knowledge previously stored in the original foundation model.To address this, we propose SAM-Med3D-MoE, a novel framework that seamlessly integrates task-specific finetuned models with the foundational model, creating a unified model at minimal additional training expense for an extra gating network. This gating network, in conjunction with a selection strategy, allows the unified model to achieve comparable performance of the original models in their respective tasks both general and specialized without updating any parameters of them.Our comprehensive experiments demonstrate the efficacy of SAM-Med3D-MoE, with an average Dice performance increase from 53 to 56.4 on 15 specific classes. It especially gets remarkable gains of 29.6, 8.5, 11.2 on the spinal cord, esophagus, and right hip, respectively. Additionally, it achieves 48.9 Dice on the challenging SPPIN2023 Challenge, significantly surpassing the general expert's performance of 32.3. We anticipate that SAM-Med3D-MoE can serve as a new framework for adapting the foundation model to specific areas in medical image analysis. Codes and datasets will be publicly available.
Motion-Informed Deep Learning for Brain MR Image Reconstruction Framework
May 28, 2024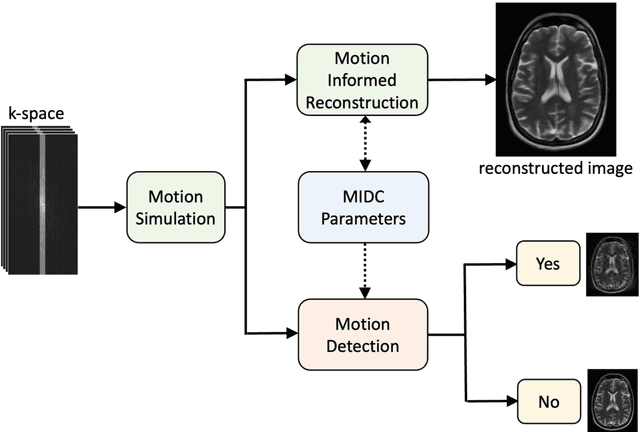
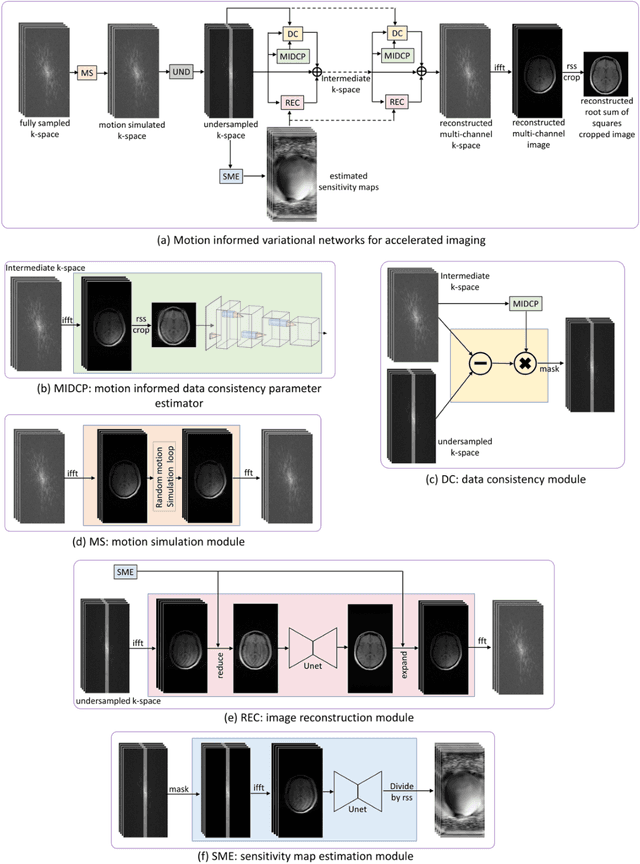
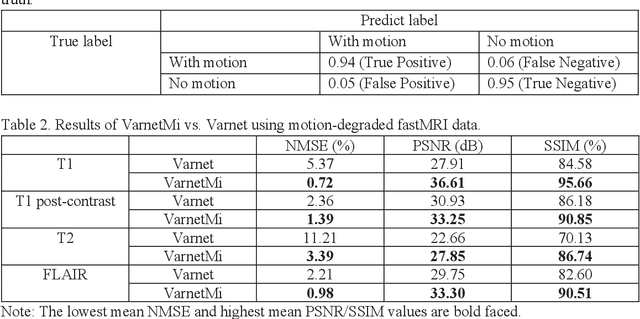
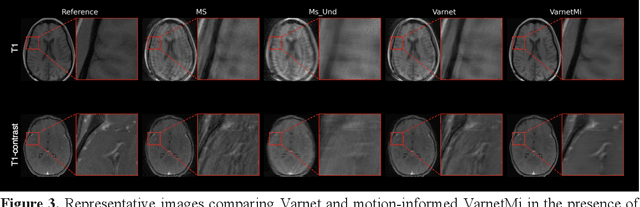
Abstract:Motion artifacts in Magnetic Resonance Imaging (MRI) are one of the frequently occurring artifacts due to patient movements during scanning. Motion is estimated to be present in approximately 30% of clinical MRI scans; however, motion has not been explicitly modeled within deep learning image reconstruction models. Deep learning (DL) algorithms have been demonstrated to be effective for both the image reconstruction task and the motion correction task, but the two tasks are considered separately. The image reconstruction task involves removing undersampling artifacts such as noise and aliasing artifacts, whereas motion correction involves removing artifacts including blurring, ghosting, and ringing. In this work, we propose a novel method to simultaneously accelerate imaging and correct motion. This is achieved by integrating a motion module into the deep learning-based MRI reconstruction process, enabling real-time detection and correction of motion. We model motion as a tightly integrated auxiliary layer in the deep learning model during training, making the deep learning model 'motion-informed'. During inference, image reconstruction is performed from undersampled raw k-space data using a trained motion-informed DL model. Experimental results demonstrate that the proposed motion-informed deep learning image reconstruction network outperformed the conventional image reconstruction network for motion-degraded MRI datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge