Mevan Ekanayake
SeCo-INR: Semantically Conditioned Implicit Neural Representations for Improved Medical Image Super-Resolution
Sep 02, 2024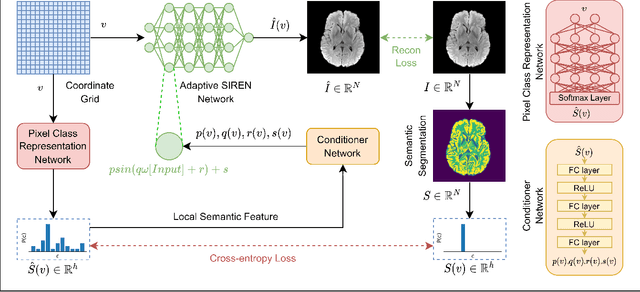
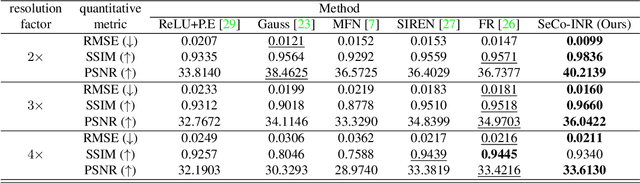
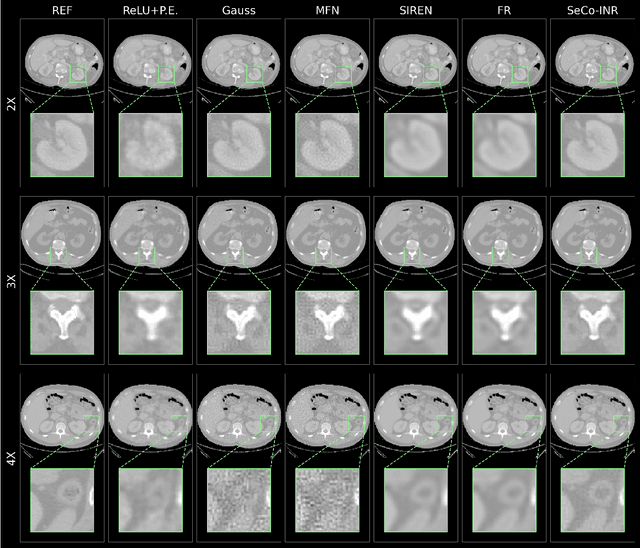
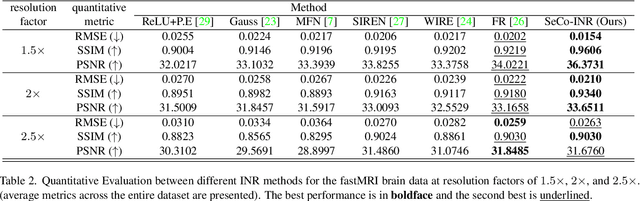
Abstract:Implicit Neural Representations (INRs) have recently advanced the field of deep learning due to their ability to learn continuous representations of signals without the need for large training datasets. Although INR methods have been studied for medical image super-resolution, their adaptability to localized priors in medical images has not been extensively explored. Medical images contain rich anatomical divisions that could provide valuable local prior information to enhance the accuracy and robustness of INRs. In this work, we propose a novel framework, referred to as the Semantically Conditioned INR (SeCo-INR), that conditions an INR using local priors from a medical image, enabling accurate model fitting and interpolation capabilities to achieve super-resolution. Our framework learns a continuous representation of the semantic segmentation features of a medical image and utilizes it to derive the optimal INR for each semantic region of the image. We tested our framework using several medical imaging modalities and achieved higher quantitative scores and more realistic super-resolution outputs compared to state-of-the-art methods.
Contrastive Learning MRI Reconstruction
Jun 01, 2023

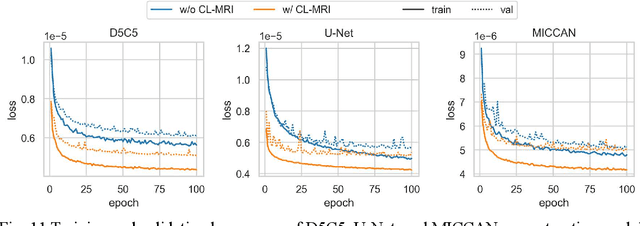

Abstract:Purpose: We propose a novel contrastive learning latent space representation for MRI datasets with partially acquired scans. We show that this latent space can be utilized for accelerated MR image reconstruction. Theory and Methods: Our novel framework, referred to as COLADA (stands for Contrastive Learning for highly accelerated MR image reconstruction), maximizes the mutual information between differently accelerated images of an MRI scan by using self-supervised contrastive learning. In other words, it attempts to "pull" the latent representations of the same scan together and "push" the latent representations of other scans away. The generated MRI latent space is subsequently utilized for MR image reconstruction and the performance was assessed in comparison to several baseline deep learning reconstruction methods. Furthermore, the quality of the proposed latent space representation was analyzed using Alignment and Uniformity. Results: COLADA comprehensively outperformed other reconstruction methods with robustness to variations in undersampling patterns, pathological abnormalities, and noise in k-space during inference. COLADA proved the high quality of reconstruction on unseen data with minimal fine-tuning. The analysis of representation quality suggests that the contrastive features produced by COLADA are optimally distributed in latent space. Conclusion: To the best of our knowledge, this is the first attempt to utilize contrastive learning on differently accelerated images for MR image reconstruction. The proposed latent space representation has practical usage due to a large number of existing partially sampled datasets. This implies the possibility of exploring self-supervised contrastive learning further to enhance the latent space of MRI for image reconstruction.
PixCUE: Joint Uncertainty Estimation and Image Reconstruction in MRI using Deep Pixel Classification
Mar 08, 2023



Abstract:Deep learning (DL) models are capable of successfully exploiting latent representations in MR data and have become state-of-the-art for accelerated MRI reconstruction. However, undersampling the measurements in k-space as well as the over- or under-parameterized and non-transparent nature of DL make these models exposed to uncertainty. Consequently, uncertainty estimation has become a major issue in DL MRI reconstruction. To estimate uncertainty, Monte Carlo (MC) inference techniques have become a common practice where multiple reconstructions are utilized to compute the variance in reconstruction as a measurement of uncertainty. However, these methods demand high computational costs as they require multiple inferences through the DL model. To this end, we introduce a method to estimate uncertainty during MRI reconstruction using a pixel classification framework. The proposed method, PixCUE (stands for Pixel Classification Uncertainty Estimation) produces the reconstructed image along with an uncertainty map during a single forward pass through the DL model. We demonstrate that this approach generates uncertainty maps that highly correlate with the reconstruction errors with respect to various MR imaging sequences and under numerous adversarial conditions. We also show that the estimated uncertainties are correlated to that of the conventional MC method. We further provide an empirical relationship between the uncertainty estimations using PixCUE and well-established reconstruction metrics such as NMSE, PSNR, and SSIM. We conclude that PixCUE is capable of reliably estimating the uncertainty in MRI reconstruction with a minimum additional computational cost.
Multi-head Cascaded Swin Transformers with Attention to k-space Sampling Pattern for Accelerated MRI Reconstruction
Jul 18, 2022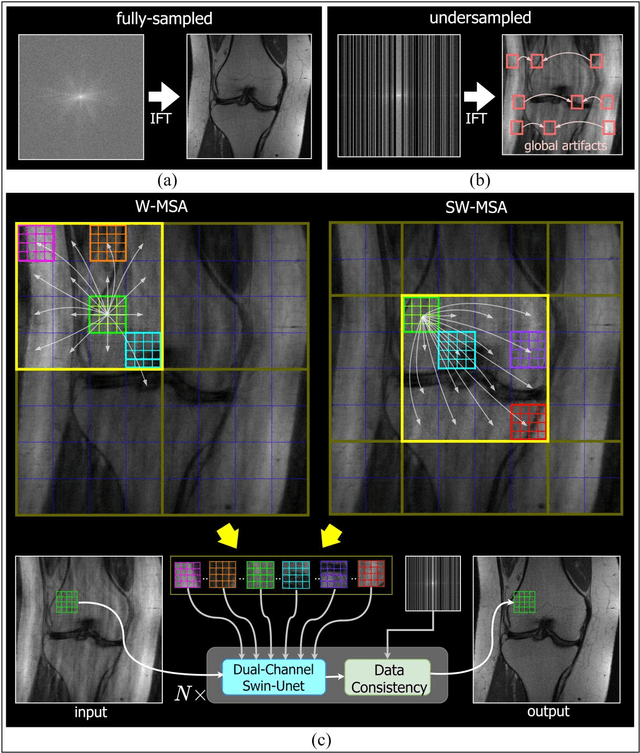
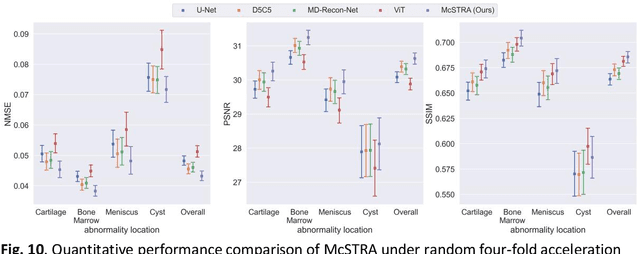
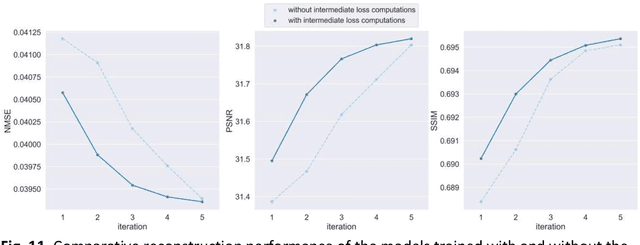

Abstract:Global correlations are widely seen in human anatomical structures due to similarity across tissues and bones. These correlations are reflected in magnetic resonance imaging (MRI) scans as a result of close-range proton density and T1/T2 parameter. Furthermore, to achieve accelerated MRI, k-space data are undersampled which causes global aliasing artifacts. Convolutional neural network (CNN) models are widely utilized for accelerated MRI reconstruction, but those models are limited in capturing global correlations due to the intrinsic locality of the convolution operation. The self-attention-based transformer models are capable of capturing global correlations among image features, however, the current contributions of transformer models for MRI reconstruction are minute. The existing contributions mostly provide CNN-transformer hybrid solutions and rarely leverage the physics of MRI. In this paper, we propose a physics-based stand-alone (convolution free) transformer model titled, the Multi-head Cascaded Swin Transformers (McSTRA) for accelerated MRI reconstruction. McSTRA combines several interconnected MRI physics-related concepts with the transformer networks: it exploits global MR features via the shifted window self-attention mechanism; it extracts MR features belonging to different spectral components separately using a multi-head setup; it iterates between intermediate de-aliasing and k-space correction via a cascaded network with data consistency in k-space and intermediate loss computations; furthermore, we propose a novel positional embedding generation mechanism to guide self-attention utilizing the point spread function corresponding to the undersampling mask. Our model significantly outperforms state-of-the-art MRI reconstruction methods both visually and quantitatively while depicting improved resolution and removal of aliasing artifacts.
Convolutional Autoencoder for Blind Hyperspectral Image Unmixing
Nov 18, 2020



Abstract:In the remote sensing context spectral unmixing is a technique to decompose a mixed pixel into two fundamental representatives: endmembers and abundances. In this paper, a novel architecture is proposed to perform blind unmixing on hyperspectral images. The proposed architecture consists of convolutional layers followed by an autoencoder. The encoder transforms the feature space produced through convolutional layers to a latent space representation. Then, from these latent characteristics the decoder reconstructs the roll-out image of the monochrome image which is at the input of the architecture; and each single-band image is fed sequentially. Experimental results on real hyperspectral data concludes that the proposed algorithm outperforms existing unmixing methods at abundance estimation and generates competitive results for endmember extraction with RMSE and SAD as the metrics, respectively.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge