Gary Egan
Segment Any Tumour: An Uncertainty-Aware Vision Foundation Model for Whole-Body Analysis
Nov 11, 2025Abstract:Prompt-driven vision foundation models, such as the Segment Anything Model, have recently demonstrated remarkable adaptability in computer vision. However, their direct application to medical imaging remains challenging due to heterogeneous tissue structures, imaging artefacts, and low-contrast boundaries, particularly in tumours and cancer primaries leading to suboptimal segmentation in ambiguous or overlapping lesion regions. Here, we present Segment Any Tumour 3D (SAT3D), a lightweight volumetric foundation model designed to enable robust and generalisable tumour segmentation across diverse medical imaging modalities. SAT3D integrates a shifted-window vision transformer for hierarchical volumetric representation with an uncertainty-aware training pipeline that explicitly incorporates uncertainty estimates as prompts to guide reliable boundary prediction in low-contrast regions. Adversarial learning further enhances model performance for the ambiguous pathological regions. We benchmark SAT3D against three recent vision foundation models and nnUNet across 11 publicly available datasets, encompassing 3,884 tumour and cancer cases for training and 694 cases for in-distribution evaluation. Trained on 17,075 3D volume-mask pairs across multiple modalities and cancer primaries, SAT3D demonstrates strong generalisation and robustness. To facilitate practical use and clinical translation, we developed a 3D Slicer plugin that enables interactive, prompt-driven segmentation and visualisation using the trained SAT3D model. Extensive experiments highlight its effectiveness in improving segmentation accuracy under challenging and out-of-distribution scenarios, underscoring its potential as a scalable foundation model for medical image analysis.
Deep kernel representations of latent space features for low-dose PET-MR imaging robust to variable dose reduction
Sep 10, 2024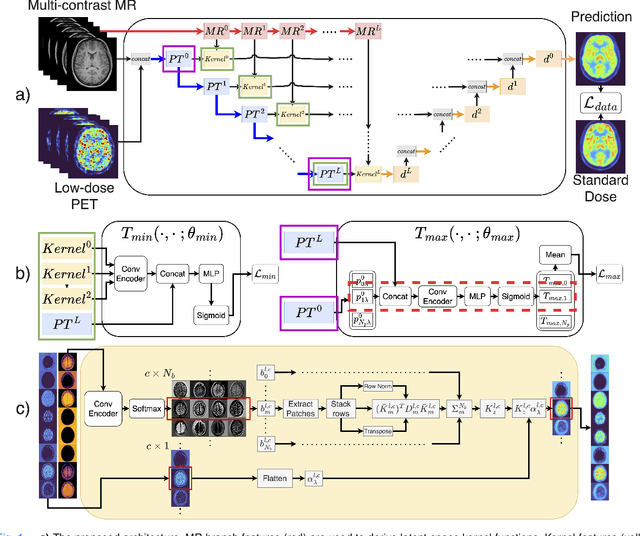
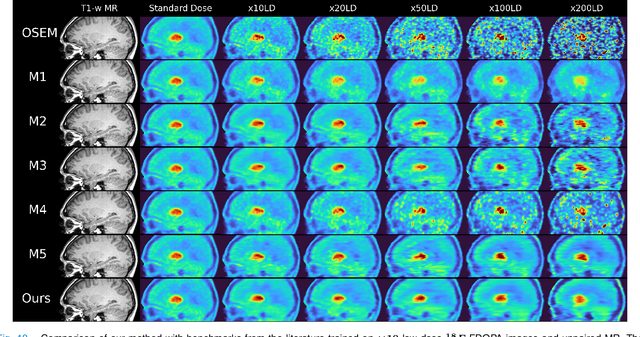
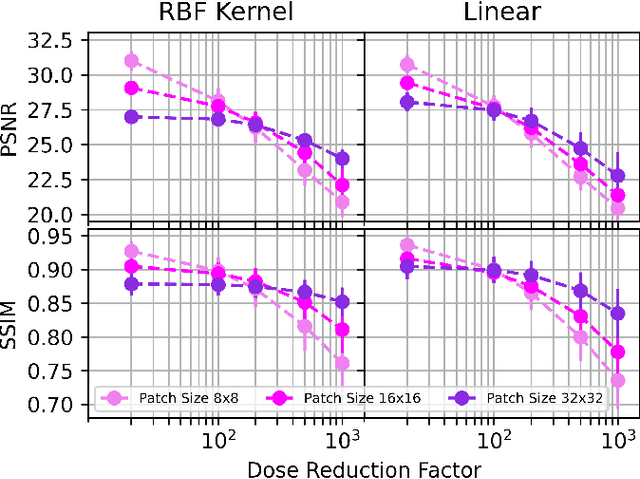
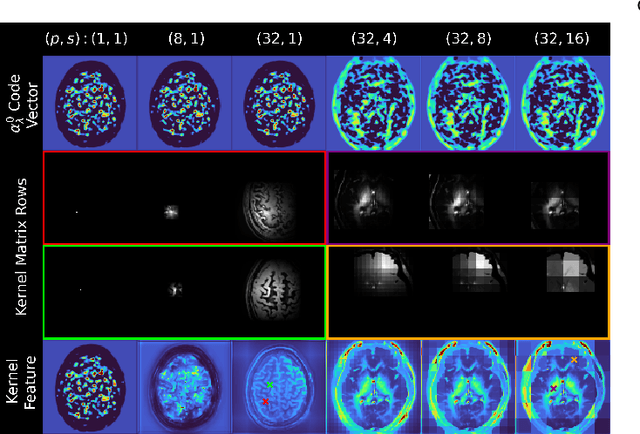
Abstract:Low-dose positron emission tomography (PET) image reconstruction methods have potential to significantly improve PET as an imaging modality. Deep learning provides a promising means of incorporating prior information into the image reconstruction problem to produce quantitatively accurate images from compromised signal. Deep learning-based methods for low-dose PET are generally poorly conditioned and perform unreliably on images with features not present in the training distribution. We present a method which explicitly models deep latent space features using a robust kernel representation, providing robust performance on previously unseen dose reduction factors. Additional constraints on the information content of deep latent features allow for tuning in-distribution accuracy and generalisability. Tests with out-of-distribution dose reduction factors ranging from $\times 10$ to $\times 1000$ and with both paired and unpaired MR, demonstrate significantly improved performance relative to conventional deep-learning methods trained using the same data. Code:https://github.com/cameronPain
SeCo-INR: Semantically Conditioned Implicit Neural Representations for Improved Medical Image Super-Resolution
Sep 02, 2024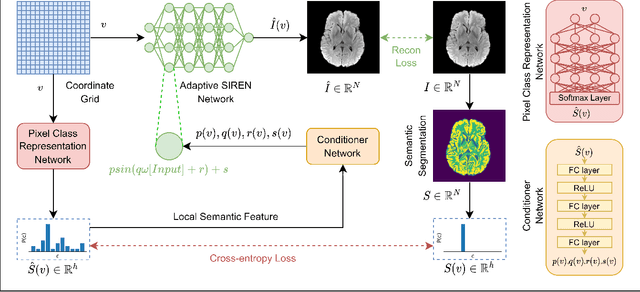
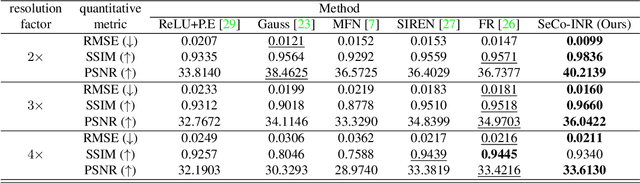
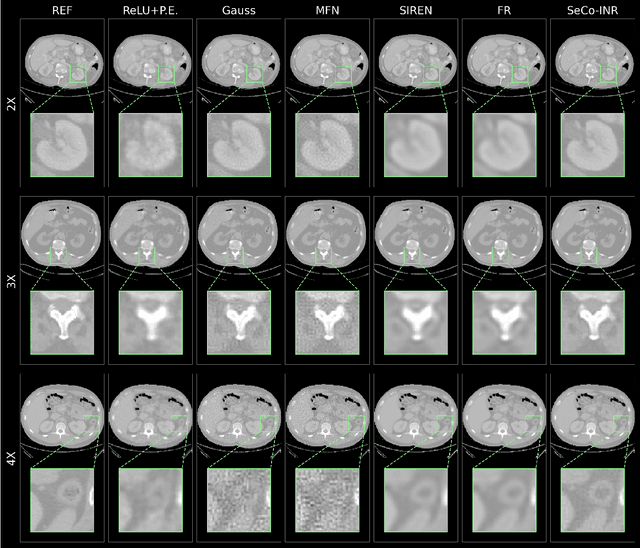
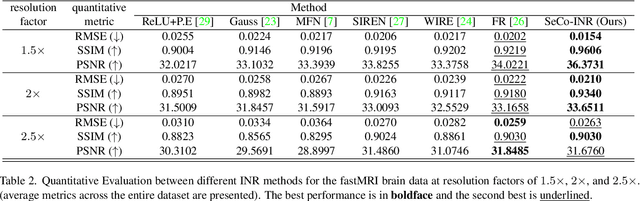
Abstract:Implicit Neural Representations (INRs) have recently advanced the field of deep learning due to their ability to learn continuous representations of signals without the need for large training datasets. Although INR methods have been studied for medical image super-resolution, their adaptability to localized priors in medical images has not been extensively explored. Medical images contain rich anatomical divisions that could provide valuable local prior information to enhance the accuracy and robustness of INRs. In this work, we propose a novel framework, referred to as the Semantically Conditioned INR (SeCo-INR), that conditions an INR using local priors from a medical image, enabling accurate model fitting and interpolation capabilities to achieve super-resolution. Our framework learns a continuous representation of the semantic segmentation features of a medical image and utilizes it to derive the optimal INR for each semantic region of the image. We tested our framework using several medical imaging modalities and achieved higher quantitative scores and more realistic super-resolution outputs compared to state-of-the-art methods.
Motion-Informed Deep Learning for Brain MR Image Reconstruction Framework
May 28, 2024
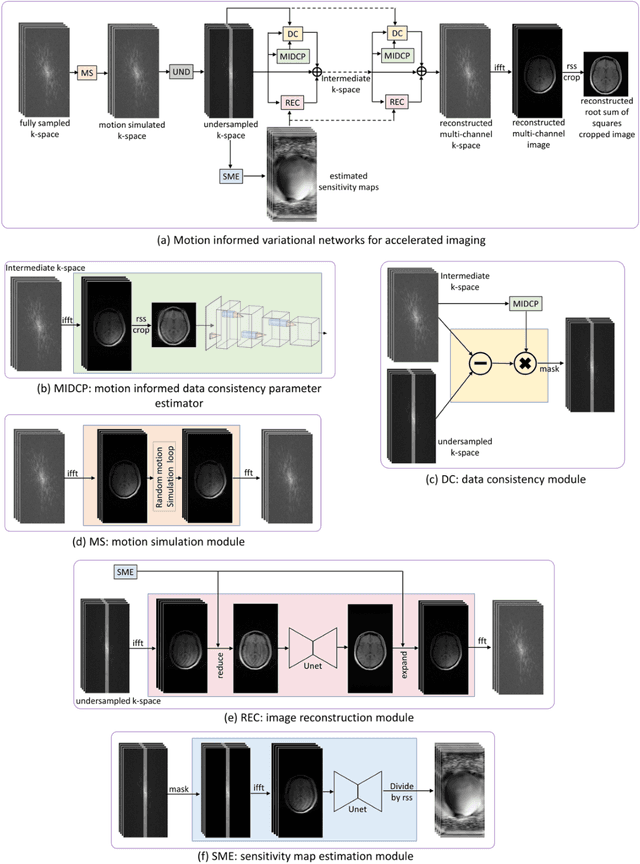
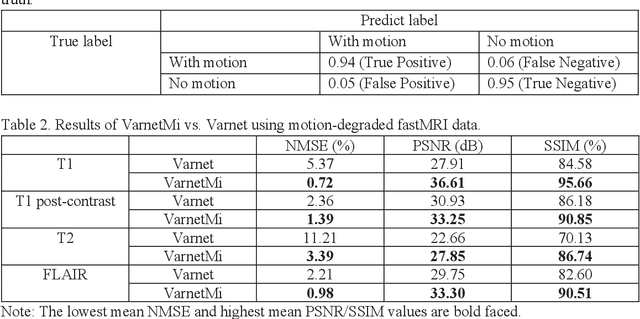
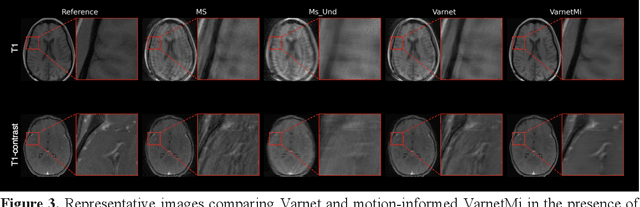
Abstract:Motion artifacts in Magnetic Resonance Imaging (MRI) are one of the frequently occurring artifacts due to patient movements during scanning. Motion is estimated to be present in approximately 30% of clinical MRI scans; however, motion has not been explicitly modeled within deep learning image reconstruction models. Deep learning (DL) algorithms have been demonstrated to be effective for both the image reconstruction task and the motion correction task, but the two tasks are considered separately. The image reconstruction task involves removing undersampling artifacts such as noise and aliasing artifacts, whereas motion correction involves removing artifacts including blurring, ghosting, and ringing. In this work, we propose a novel method to simultaneously accelerate imaging and correct motion. This is achieved by integrating a motion module into the deep learning-based MRI reconstruction process, enabling real-time detection and correction of motion. We model motion as a tightly integrated auxiliary layer in the deep learning model during training, making the deep learning model 'motion-informed'. During inference, image reconstruction is performed from undersampled raw k-space data using a trained motion-informed DL model. Experimental results demonstrate that the proposed motion-informed deep learning image reconstruction network outperformed the conventional image reconstruction network for motion-degraded MRI datasets.
Contrastive Learning MRI Reconstruction
Jun 01, 2023

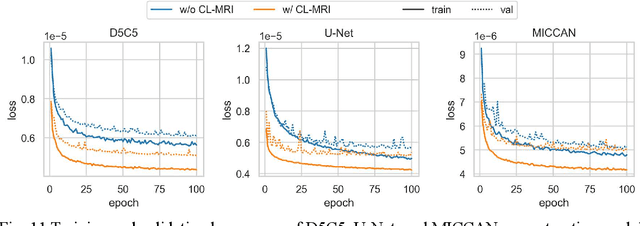

Abstract:Purpose: We propose a novel contrastive learning latent space representation for MRI datasets with partially acquired scans. We show that this latent space can be utilized for accelerated MR image reconstruction. Theory and Methods: Our novel framework, referred to as COLADA (stands for Contrastive Learning for highly accelerated MR image reconstruction), maximizes the mutual information between differently accelerated images of an MRI scan by using self-supervised contrastive learning. In other words, it attempts to "pull" the latent representations of the same scan together and "push" the latent representations of other scans away. The generated MRI latent space is subsequently utilized for MR image reconstruction and the performance was assessed in comparison to several baseline deep learning reconstruction methods. Furthermore, the quality of the proposed latent space representation was analyzed using Alignment and Uniformity. Results: COLADA comprehensively outperformed other reconstruction methods with robustness to variations in undersampling patterns, pathological abnormalities, and noise in k-space during inference. COLADA proved the high quality of reconstruction on unseen data with minimal fine-tuning. The analysis of representation quality suggests that the contrastive features produced by COLADA are optimally distributed in latent space. Conclusion: To the best of our knowledge, this is the first attempt to utilize contrastive learning on differently accelerated images for MR image reconstruction. The proposed latent space representation has practical usage due to a large number of existing partially sampled datasets. This implies the possibility of exploring self-supervised contrastive learning further to enhance the latent space of MRI for image reconstruction.
PixCUE: Joint Uncertainty Estimation and Image Reconstruction in MRI using Deep Pixel Classification
Mar 08, 2023



Abstract:Deep learning (DL) models are capable of successfully exploiting latent representations in MR data and have become state-of-the-art for accelerated MRI reconstruction. However, undersampling the measurements in k-space as well as the over- or under-parameterized and non-transparent nature of DL make these models exposed to uncertainty. Consequently, uncertainty estimation has become a major issue in DL MRI reconstruction. To estimate uncertainty, Monte Carlo (MC) inference techniques have become a common practice where multiple reconstructions are utilized to compute the variance in reconstruction as a measurement of uncertainty. However, these methods demand high computational costs as they require multiple inferences through the DL model. To this end, we introduce a method to estimate uncertainty during MRI reconstruction using a pixel classification framework. The proposed method, PixCUE (stands for Pixel Classification Uncertainty Estimation) produces the reconstructed image along with an uncertainty map during a single forward pass through the DL model. We demonstrate that this approach generates uncertainty maps that highly correlate with the reconstruction errors with respect to various MR imaging sequences and under numerous adversarial conditions. We also show that the estimated uncertainties are correlated to that of the conventional MC method. We further provide an empirical relationship between the uncertainty estimations using PixCUE and well-established reconstruction metrics such as NMSE, PSNR, and SSIM. We conclude that PixCUE is capable of reliably estimating the uncertainty in MRI reconstruction with a minimum additional computational cost.
Hybrid Window Attention Based Transformer Architecture for Brain Tumor Segmentation
Sep 16, 2022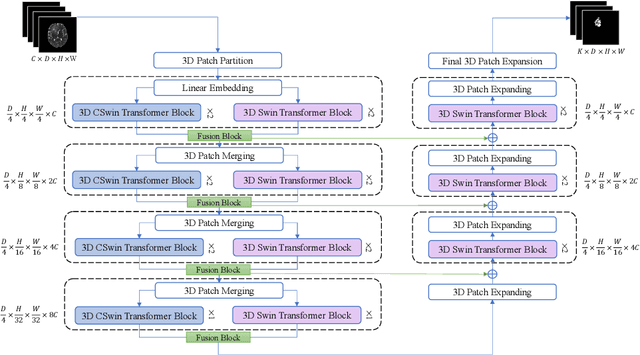
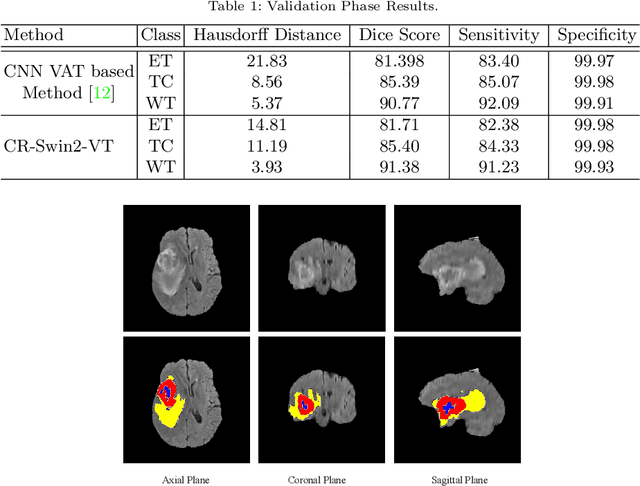
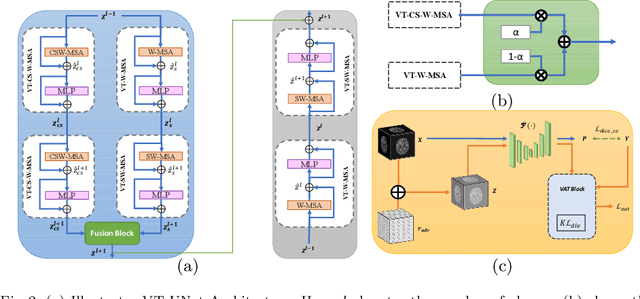
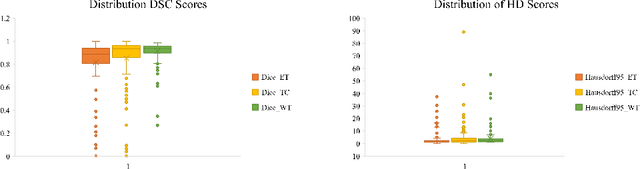
Abstract:As intensities of MRI volumes are inconsistent across institutes, it is essential to extract universal features of multi-modal MRIs to precisely segment brain tumors. In this concept, we propose a volumetric vision transformer that follows two windowing strategies in attention for extracting fine features and local distributional smoothness (LDS) during model training inspired by virtual adversarial training (VAT) to make the model robust. We trained and evaluated network architecture on the FeTS Challenge 2022 dataset. Our performance on the online validation dataset is as follows: Dice Similarity Score of 81.71%, 91.38% and 85.40%; Hausdorff Distance (95%) of 14.81 mm, 3.93 mm, 11.18 mm for the enhancing tumor, whole tumor, and tumor core, respectively. Overall, the experimental results verify our method's effectiveness by yielding better performance in segmentation accuracy for each tumor sub-region. Our code implementation is publicly available : https://github.com/himashi92/vizviva_fets_2022
Multi-head Cascaded Swin Transformers with Attention to k-space Sampling Pattern for Accelerated MRI Reconstruction
Jul 18, 2022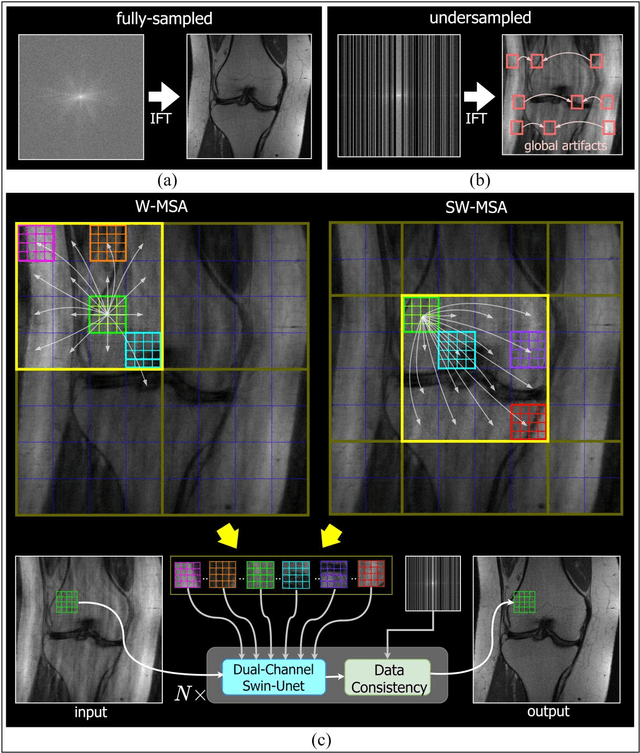
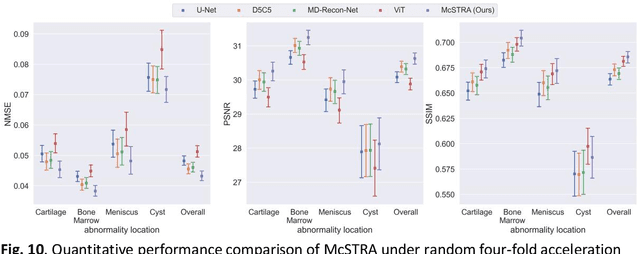
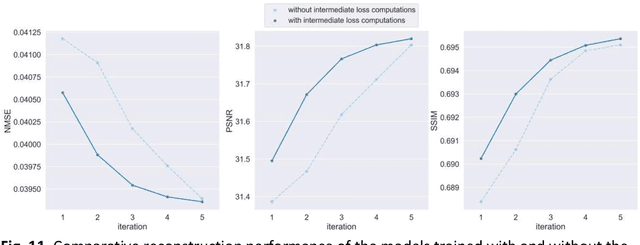

Abstract:Global correlations are widely seen in human anatomical structures due to similarity across tissues and bones. These correlations are reflected in magnetic resonance imaging (MRI) scans as a result of close-range proton density and T1/T2 parameter. Furthermore, to achieve accelerated MRI, k-space data are undersampled which causes global aliasing artifacts. Convolutional neural network (CNN) models are widely utilized for accelerated MRI reconstruction, but those models are limited in capturing global correlations due to the intrinsic locality of the convolution operation. The self-attention-based transformer models are capable of capturing global correlations among image features, however, the current contributions of transformer models for MRI reconstruction are minute. The existing contributions mostly provide CNN-transformer hybrid solutions and rarely leverage the physics of MRI. In this paper, we propose a physics-based stand-alone (convolution free) transformer model titled, the Multi-head Cascaded Swin Transformers (McSTRA) for accelerated MRI reconstruction. McSTRA combines several interconnected MRI physics-related concepts with the transformer networks: it exploits global MR features via the shifted window self-attention mechanism; it extracts MR features belonging to different spectral components separately using a multi-head setup; it iterates between intermediate de-aliasing and k-space correction via a cascaded network with data consistency in k-space and intermediate loss computations; furthermore, we propose a novel positional embedding generation mechanism to guide self-attention utilizing the point spread function corresponding to the undersampling mask. Our model significantly outperforms state-of-the-art MRI reconstruction methods both visually and quantitatively while depicting improved resolution and removal of aliasing artifacts.
Reciprocal Adversarial Learning for Brain Tumor Segmentation: A Solution to BraTS Challenge 2021 Segmentation Task
Jan 11, 2022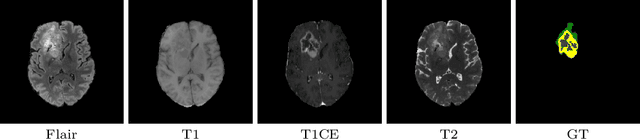
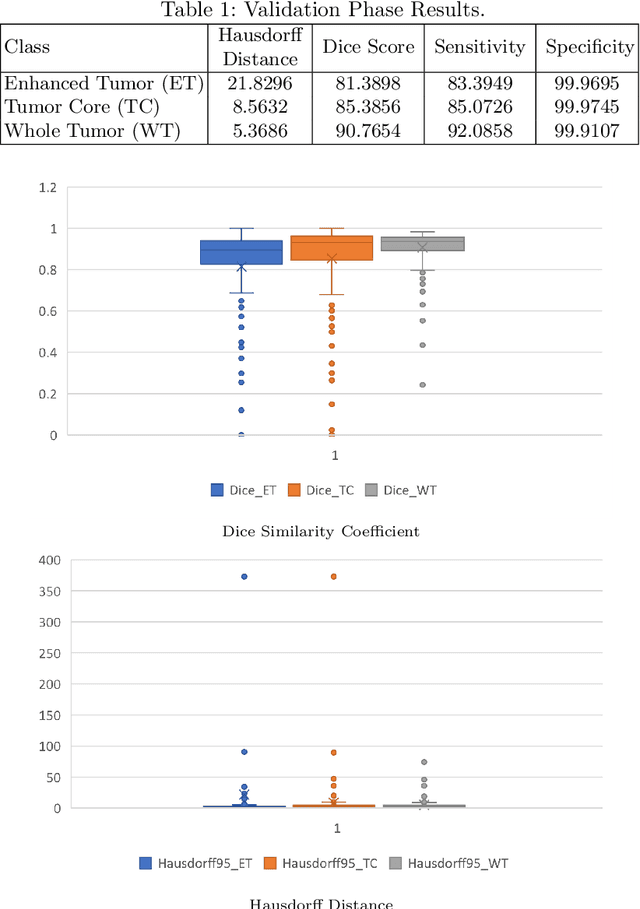
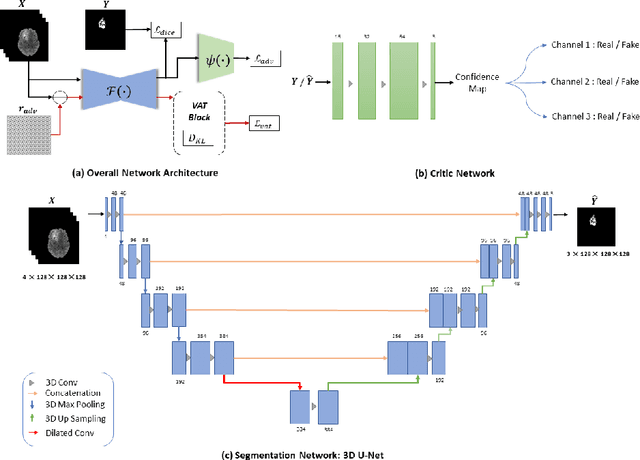
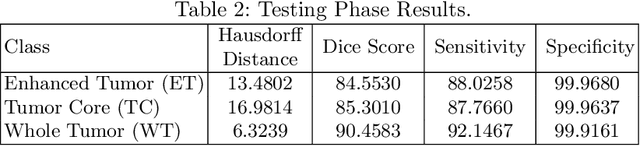
Abstract:This paper proposes an adversarial learning based training approach for brain tumor segmentation task. In this concept, the 3D segmentation network learns from dual reciprocal adversarial learning approaches. To enhance the generalization across the segmentation predictions and to make the segmentation network robust, we adhere to the Virtual Adversarial Training approach by generating more adversarial examples via adding some noise on original patient data. By incorporating a critic that acts as a quantitative subjective referee, the segmentation network learns from the uncertainty information associated with segmentation results. We trained and evaluated network architecture on the RSNA-ASNR-MICCAI BraTS 2021 dataset. Our performance on the online validation dataset is as follows: Dice Similarity Score of 81.38%, 90.77% and 85.39%; Hausdorff Distance (95\%) of 21.83 mm, 5.37 mm, 8.56 mm for the enhancing tumor, whole tumor and tumor core, respectively. Similarly, our approach achieved a Dice Similarity Score of 84.55%, 90.46% and 85.30%, as well as Hausdorff Distance (95\%) of 13.48 mm, 6.32 mm and 16.98 mm on the final test dataset. Overall, our proposed approach yielded better performance in segmentation accuracy for each tumor sub-region. Our code implementation is publicly available at https://github.com/himashi92/vizviva_brats_2021
A Volumetric Transformer for Accurate 3D Tumor Segmentation
Nov 26, 2021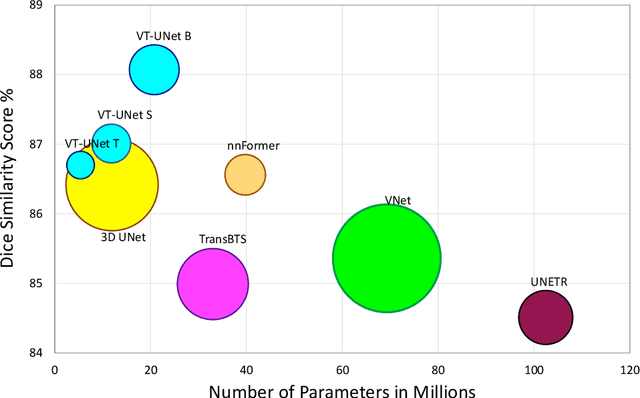
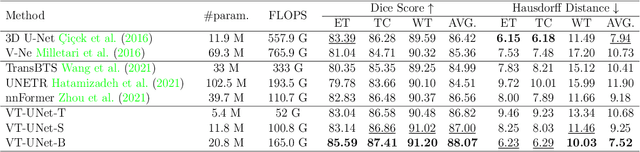
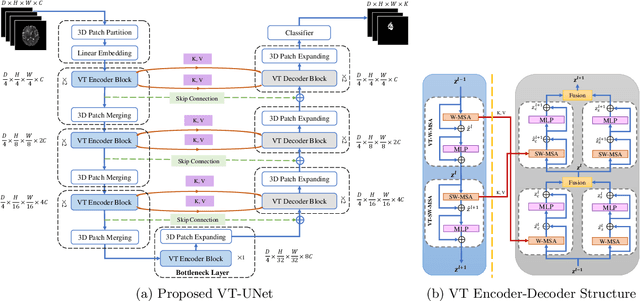
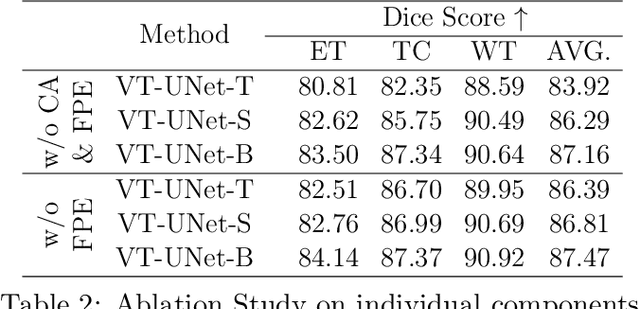
Abstract:This paper presents a Transformer architecture for volumetric medical image segmentation. Designing a computationally efficient Transformer architecture for volumetric segmentation is a challenging task. It requires keeping a complex balance in encoding local and global spatial cues, and preserving information along all axes of the volumetric data. The proposed volumetric Transformer has a U-shaped encoder-decoder design that processes the input voxels in their entirety. Our encoder has two consecutive self-attention layers to simultaneously encode local and global cues, and our decoder has novel parallel shifted window based self and cross attention blocks to capture fine details for boundary refinement by subsuming Fourier position encoding. Our proposed design choices result in a computationally efficient architecture, which demonstrates promising results on Brain Tumor Segmentation (BraTS) 2021, and Medical Segmentation Decathlon (Pancreas and Liver) datasets for tumor segmentation. We further show that the representations learned by our model transfer better across-datasets and are robust against data corruptions. \href{https://github.com/himashi92/VT-UNet}{Our code implementation is publicly available}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge