Meng Law
Segment Any Tumour: An Uncertainty-Aware Vision Foundation Model for Whole-Body Analysis
Nov 11, 2025Abstract:Prompt-driven vision foundation models, such as the Segment Anything Model, have recently demonstrated remarkable adaptability in computer vision. However, their direct application to medical imaging remains challenging due to heterogeneous tissue structures, imaging artefacts, and low-contrast boundaries, particularly in tumours and cancer primaries leading to suboptimal segmentation in ambiguous or overlapping lesion regions. Here, we present Segment Any Tumour 3D (SAT3D), a lightweight volumetric foundation model designed to enable robust and generalisable tumour segmentation across diverse medical imaging modalities. SAT3D integrates a shifted-window vision transformer for hierarchical volumetric representation with an uncertainty-aware training pipeline that explicitly incorporates uncertainty estimates as prompts to guide reliable boundary prediction in low-contrast regions. Adversarial learning further enhances model performance for the ambiguous pathological regions. We benchmark SAT3D against three recent vision foundation models and nnUNet across 11 publicly available datasets, encompassing 3,884 tumour and cancer cases for training and 694 cases for in-distribution evaluation. Trained on 17,075 3D volume-mask pairs across multiple modalities and cancer primaries, SAT3D demonstrates strong generalisation and robustness. To facilitate practical use and clinical translation, we developed a 3D Slicer plugin that enables interactive, prompt-driven segmentation and visualisation using the trained SAT3D model. Extensive experiments highlight its effectiveness in improving segmentation accuracy under challenging and out-of-distribution scenarios, underscoring its potential as a scalable foundation model for medical image analysis.
McCaD: Multi-Contrast MRI Conditioned, Adaptive Adversarial Diffusion Model for High-Fidelity MRI Synthesis
Sep 01, 2024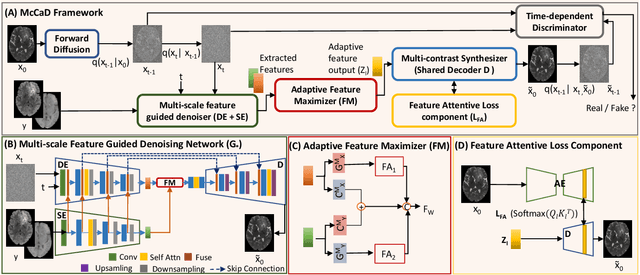



Abstract:Magnetic Resonance Imaging (MRI) is instrumental in clinical diagnosis, offering diverse contrasts that provide comprehensive diagnostic information. However, acquiring multiple MRI contrasts is often constrained by high costs, long scanning durations, and patient discomfort. Current synthesis methods, typically focused on single-image contrasts, fall short in capturing the collective nuances across various contrasts. Moreover, existing methods for multi-contrast MRI synthesis often fail to accurately map feature-level information across multiple imaging contrasts. We introduce McCaD (Multi-Contrast MRI Conditioned Adaptive Adversarial Diffusion), a novel framework leveraging an adversarial diffusion model conditioned on multiple contrasts for high-fidelity MRI synthesis. McCaD significantly enhances synthesis accuracy by employing a multi-scale, feature-guided mechanism, incorporating denoising and semantic encoders. An adaptive feature maximization strategy and a spatial feature-attentive loss have been introduced to capture more intrinsic features across multiple contrasts. This facilitates a precise and comprehensive feature-guided denoising process. Extensive experiments on tumor and healthy multi-contrast MRI datasets demonstrated that the McCaD outperforms state-of-the-art baselines quantitively and qualitatively. The code is provided with supplementary materials.
The RSNA Abdominal Traumatic Injury CT (RATIC) Dataset
May 30, 2024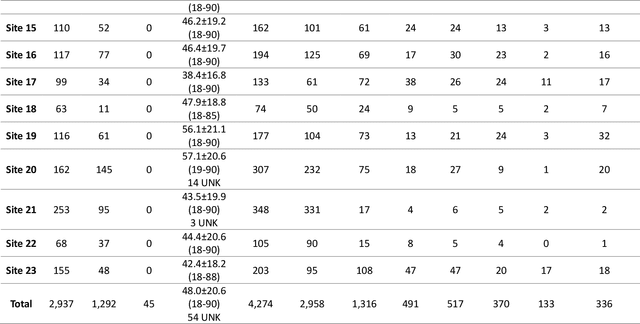
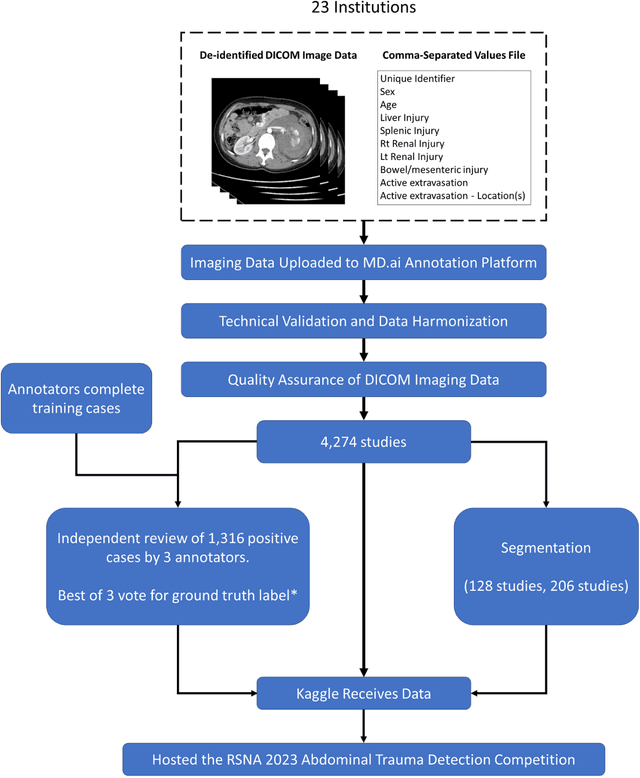
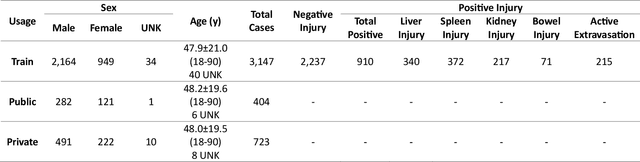
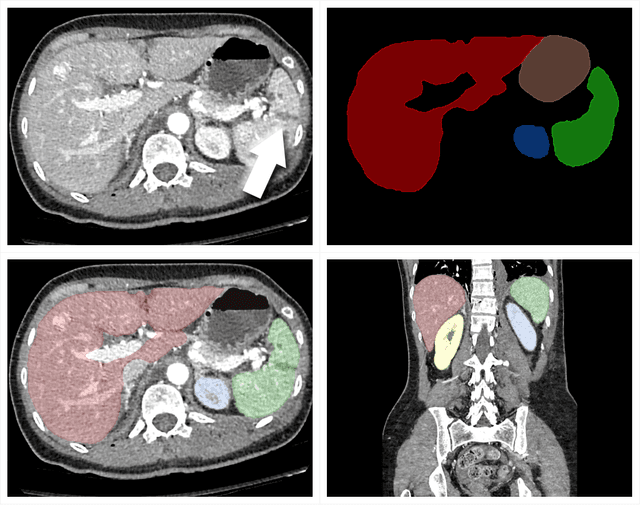
Abstract:The RSNA Abdominal Traumatic Injury CT (RATIC) dataset is the largest publicly available collection of adult abdominal CT studies annotated for traumatic injuries. This dataset includes 4,274 studies from 23 institutions across 14 countries. The dataset is freely available for non-commercial use via Kaggle at https://www.kaggle.com/competitions/rsna-2023-abdominal-trauma-detection. Created for the RSNA 2023 Abdominal Trauma Detection competition, the dataset encourages the development of advanced machine learning models for detecting abdominal injuries on CT scans. The dataset encompasses detection and classification of traumatic injuries across multiple organs, including the liver, spleen, kidneys, bowel, and mesentery. Annotations were created by expert radiologists from the American Society of Emergency Radiology (ASER) and Society of Abdominal Radiology (SAR). The dataset is annotated at multiple levels, including the presence of injuries in three solid organs with injury grading, image-level annotations for active extravasations and bowel injury, and voxelwise segmentations of each of the potentially injured organs. With the release of this dataset, we hope to facilitate research and development in machine learning and abdominal trauma that can lead to improved patient care and outcomes.
Perivascular space Identification Nnunet for Generalised Usage (PINGU)
May 17, 2024Abstract:Perivascular spaces(PVSs) form a central component of the brain\'s waste clearance system, the glymphatic system. These structures are visible on MRI images, and their morphology is associated with aging and neurological disease. Manual quantification of PVS is time consuming and subjective. Numerous deep learning methods for PVS segmentation have been developed, however the majority have been developed and evaluated on homogenous datasets and high resolution scans, perhaps limiting their applicability for the wide range of image qualities acquired in clinic and research. In this work we train a nnUNet, a top-performing biomedical image segmentation algorithm, on a heterogenous training sample of manually segmented MRI images of a range of different qualities and resolutions from 6 different datasets. These are compared to publicly available deep learning methods for 3D segmentation of PVS. The resulting model, PINGU (Perivascular space Identification Nnunet for Generalised Usage), achieved voxel and cluster level dice scores of 0.50(SD=0.15), 0.63(0.17) in the white matter(WM), and 0.54(0.11), 0.66(0.17) in the basal ganglia(BG). Performance on data from unseen sites was substantially lower for both PINGU(0.20-0.38(WM, voxel), 0.29-0.58(WM, cluster), 0.22-0.36(BG, voxel), 0.46-0.60(BG, cluster)) and the publicly available algorithms(0.18-0.30(WM, voxel), 0.29-0.38(WM cluster), 0.10-0.20(BG, voxel), 0.15-0.37(BG, cluster)), but PINGU strongly outperformed the publicly available algorithms, particularly in the BG. Finally, training PINGU on manual segmentations from a single site with homogenous scan properties gave marginally lower performances on internal cross-validation, but in some cases gave higher performance on external validation. PINGU stands out as broad-use PVS segmentation tool, with particular strength in the BG, an area of PVS related to vascular disease and pathology.
Adversarial Pulmonary Pathology Translation for Pairwise Chest X-ray Data Augmentation
Oct 11, 2019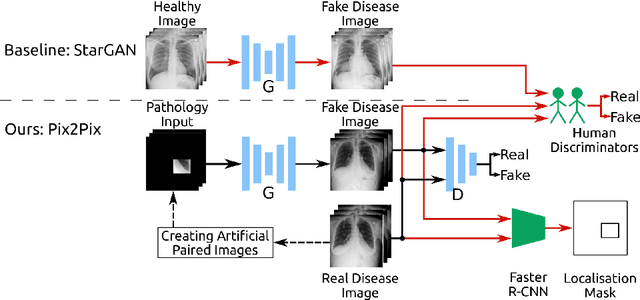
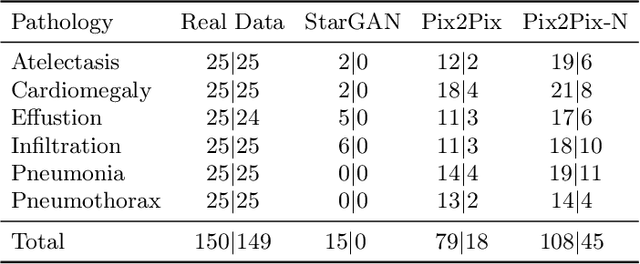
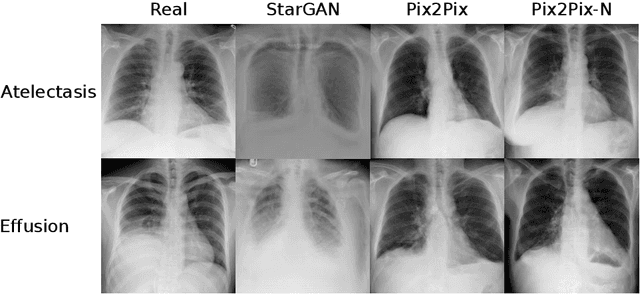

Abstract:Recent works show that Generative Adversarial Networks (GANs) can be successfully applied to chest X-ray data augmentation for lung disease recognition. However, the implausible and distorted pathology features generated from the less than perfect generator may lead to wrong clinical decisions. Why not keep the original pathology region? We proposed a novel approach that allows our generative model to generate high quality plausible images that contain undistorted pathology areas. The main idea is to design a training scheme based on an image-to-image translation network to introduce variations of new lung features around the pathology ground-truth area. Moreover, our model is able to leverage both annotated disease images and unannotated healthy lung images for the purpose of generation. We demonstrate the effectiveness of our model on two tasks: (i) we invite certified radiologists to assess the quality of the generated synthetic images against real and other state-of-the-art generative models, and (ii) data augmentation to improve the performance of disease localisation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge