Luciano M. Prevedello
for the Alzheimer's Disease Neuroimaging Initiative
The RSNA Lumbar Degenerative Imaging Spine Classification (LumbarDISC) Dataset
Jun 10, 2025Abstract:The Radiological Society of North America (RSNA) Lumbar Degenerative Imaging Spine Classification (LumbarDISC) dataset is the largest publicly available dataset of adult MRI lumbar spine examinations annotated for degenerative changes. The dataset includes 2,697 patients with a total of 8,593 image series from 8 institutions across 6 countries and 5 continents. The dataset is available for free for non-commercial use via Kaggle and RSNA Medical Imaging Resource of AI (MIRA). The dataset was created for the RSNA 2024 Lumbar Spine Degenerative Classification competition where competitors developed deep learning models to grade degenerative changes in the lumbar spine. The degree of spinal canal, subarticular recess, and neural foraminal stenosis was graded at each intervertebral disc level in the lumbar spine. The images were annotated by expert volunteer neuroradiologists and musculoskeletal radiologists from the RSNA, American Society of Neuroradiology, and the American Society of Spine Radiology. This dataset aims to facilitate research and development in machine learning and lumbar spine imaging to lead to improved patient care and clinical efficiency.
The RSNA Abdominal Traumatic Injury CT (RATIC) Dataset
May 30, 2024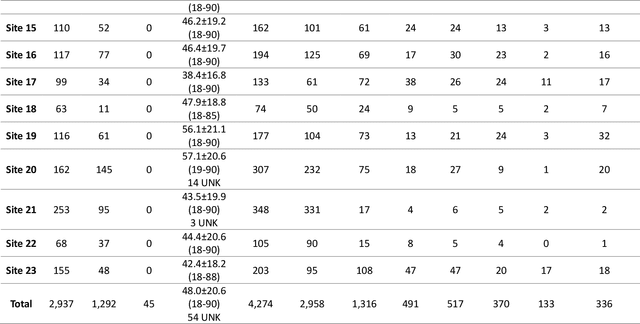
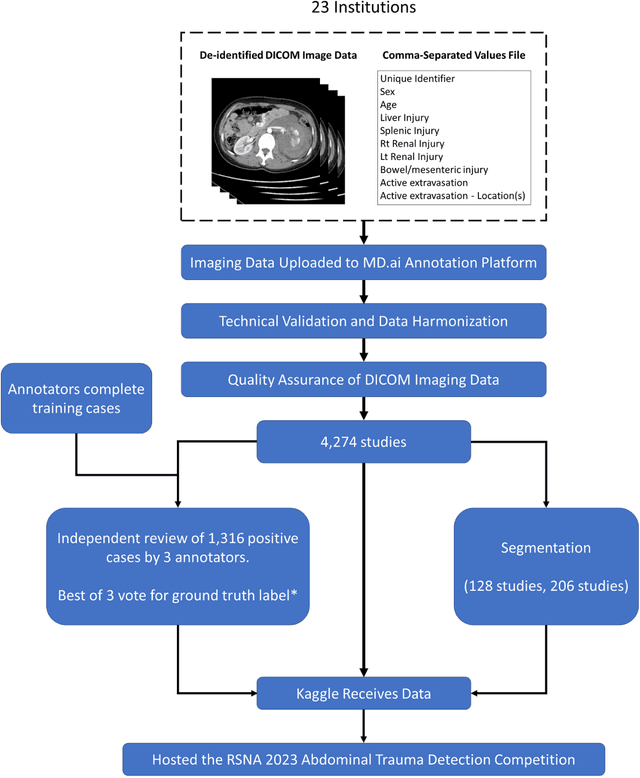
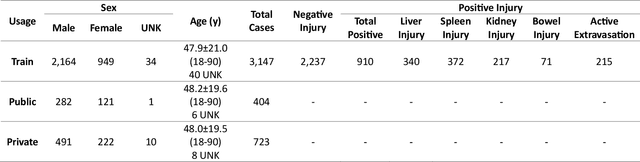
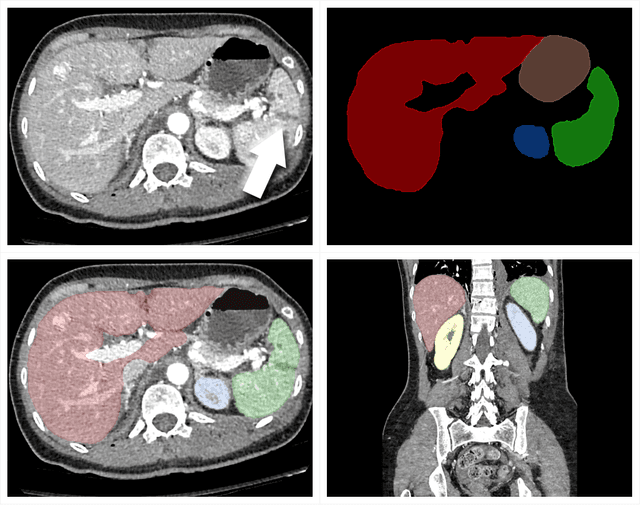
Abstract:The RSNA Abdominal Traumatic Injury CT (RATIC) dataset is the largest publicly available collection of adult abdominal CT studies annotated for traumatic injuries. This dataset includes 4,274 studies from 23 institutions across 14 countries. The dataset is freely available for non-commercial use via Kaggle at https://www.kaggle.com/competitions/rsna-2023-abdominal-trauma-detection. Created for the RSNA 2023 Abdominal Trauma Detection competition, the dataset encourages the development of advanced machine learning models for detecting abdominal injuries on CT scans. The dataset encompasses detection and classification of traumatic injuries across multiple organs, including the liver, spleen, kidneys, bowel, and mesentery. Annotations were created by expert radiologists from the American Society of Emergency Radiology (ASER) and Society of Abdominal Radiology (SAR). The dataset is annotated at multiple levels, including the presence of injuries in three solid organs with injury grading, image-level annotations for active extravasations and bowel injury, and voxelwise segmentations of each of the potentially injured organs. With the release of this dataset, we hope to facilitate research and development in machine learning and abdominal trauma that can lead to improved patient care and outcomes.
Prediction of Model Generalizability for Unseen Data: Methodology and Case Study in Brain Metastases Detection in T1-Weighted Contrast-Enhanced 3D MRI
Dec 15, 2022Abstract:A medical AI system's generalizability describes the continuity of its performance acquired from varying geographic, historical, and methodologic settings. Previous literature on this topic has mostly focused on "how" to achieve high generalizability with limited success. Instead, we aim to understand "when" the generalizability is achieved: Our study presents a medical AI system that could estimate its generalizability status for unseen data on-the-fly. We introduce a latent space mapping (LSM) approach utilizing Frechet distance loss to force the underlying training data distribution into a multivariate normal distribution. During the deployment, a given test data's LSM distribution is processed to detect its deviation from the forced distribution; hence, the AI system could predict its generalizability status for any previously unseen data set. If low model generalizability is detected, then the user is informed by a warning message. While the approach is applicable for most classification deep neural networks, we demonstrate its application to a brain metastases (BM) detector for T1-weighted contrast-enhanced (T1c) 3D MRI. The BM detection model was trained using 175 T1c studies acquired internally, and tested using (1) 42 internally and (2) 72 externally acquired exams from the publicly distributed Brain Mets dataset provided by the Stanford University School of Medicine. Generalizability scores, false positive (FP) rates, and sensitivities of the BM detector were computed for the test datasets. The model predicted its generalizability to be low for 31% of the testing data, where it produced (1) ~13.5 FPs at 76.1% BM detection sensitivity for the low and (2) ~10.5 FPs at 89.2% BM detection sensitivity for the high generalizability groups respectively. The results suggest that the proposed formulation enables a model to predict its generalizability for unseen data.
Advancing Brain Metastases Detection in T1-Weighted Contrast-Enhanced 3D MRI using Noisy Student-based Training
Nov 19, 2021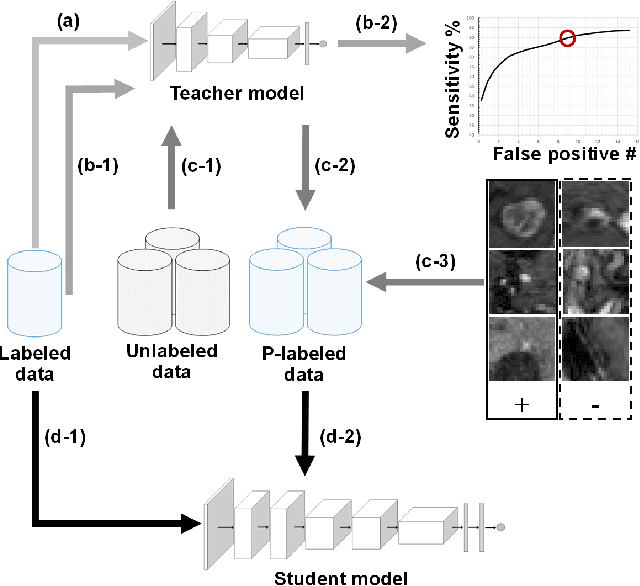
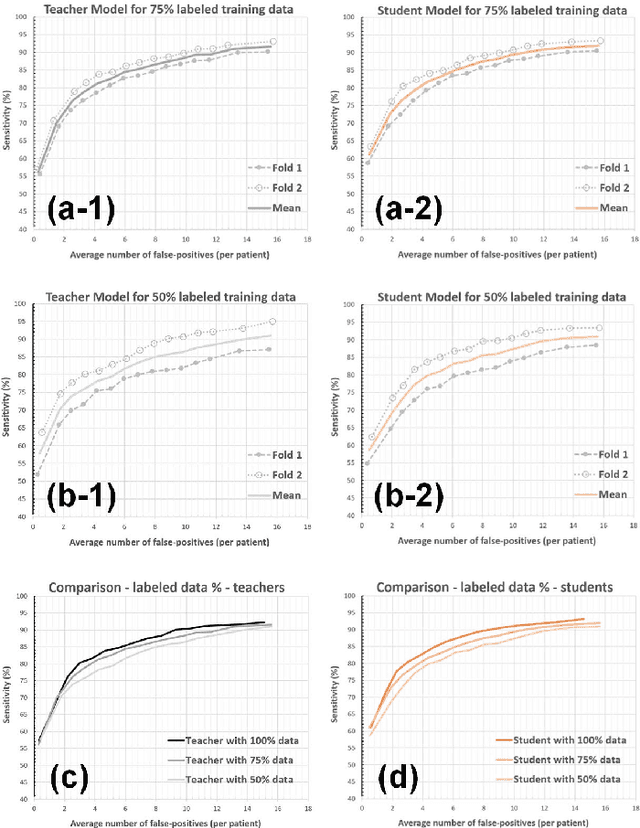
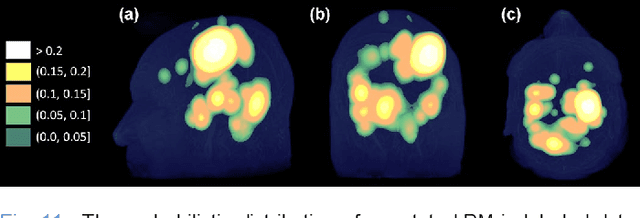
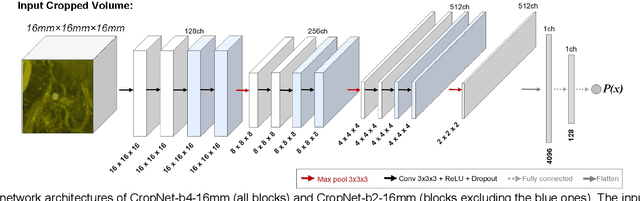
Abstract:The detection of brain metastases (BM) in their early stages could have a positive impact on the outcome of cancer patients. We previously developed a framework for detecting small BM (with diameters of less than 15mm) in T1-weighted Contrast-Enhanced 3D Magnetic Resonance images (T1c) to assist medical experts in this time-sensitive and high-stakes task. The framework utilizes a dedicated convolutional neural network (CNN) trained using labeled T1c data, where the ground truth BM segmentations were provided by a radiologist. This study aims to advance the framework with a noisy student-based self-training strategy to make use of a large corpus of unlabeled T1c data (i.e., data without BM segmentations or detections). Accordingly, the work (1) describes the student and teacher CNN architectures, (2) presents data and model noising mechanisms, and (3) introduces a novel pseudo-labeling strategy factoring in the learned BM detection sensitivity of the framework. Finally, it describes a semi-supervised learning strategy utilizing these components. We performed the validation using 217 labeled and 1247 unlabeled T1c exams via 2-fold cross-validation. The framework utilizing only the labeled exams produced 9.23 false positives for 90% BM detection sensitivity; whereas, the framework using the introduced learning strategy led to ~9% reduction in false detections (i.e., 8.44) for the same sensitivity level. Furthermore, while experiments utilizing 75% and 50% of the labeled datasets resulted in algorithm performance degradation (12.19 and 13.89 false positives respectively), the impact was less pronounced with the noisy student-based training strategy (10.79 and 12.37 false positives respectively).
The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification
Jul 05, 2021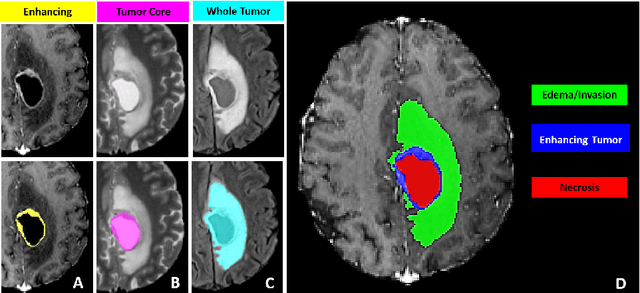
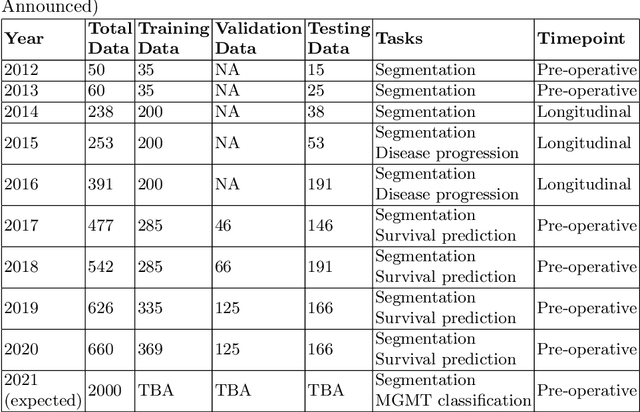
Abstract:The BraTS 2021 challenge celebrates its 10th anniversary and is jointly organized by the Radiological Society of North America (RSNA), the American Society of Neuroradiology (ASNR), and the Medical Image Computing and Computer Assisted Interventions (MICCAI) society. Since its inception, BraTS has been focusing on being a common benchmarking venue for brain glioma segmentation algorithms, with well-curated multi-institutional multi-parametric magnetic resonance imaging (mpMRI) data. Gliomas are the most common primary malignancies of the central nervous system, with varying degrees of aggressiveness and prognosis. The RSNA-ASNR-MICCAI BraTS 2021 challenge targets the evaluation of computational algorithms assessing the same tumor compartmentalization, as well as the underlying tumor's molecular characterization, in pre-operative baseline mpMRI data from 2,000 patients. Specifically, the two tasks that BraTS 2021 focuses on are: a) the segmentation of the histologically distinct brain tumor sub-regions, and b) the classification of the tumor's O[6]-methylguanine-DNA methyltransferase (MGMT) promoter methylation status. The performance evaluation of all participating algorithms in BraTS 2021 will be conducted through the Sage Bionetworks Synapse platform (Task 1) and Kaggle (Task 2), concluding in distributing to the top ranked participants monetary awards of $60,000 collectively.
Augmented Networks for Faster Brain Metastases Detection in T1-Weighted Contrast-Enhanced 3D MRI
May 27, 2021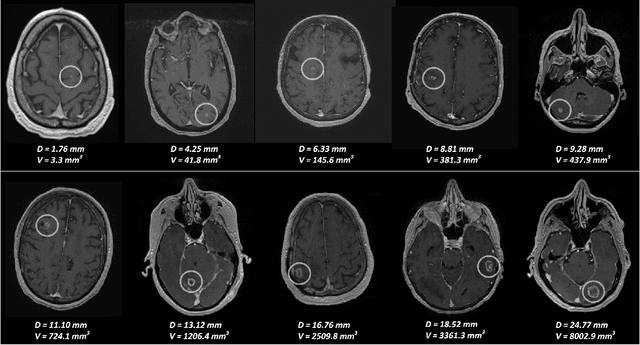
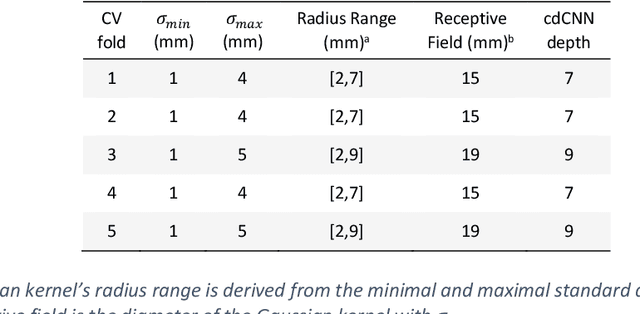
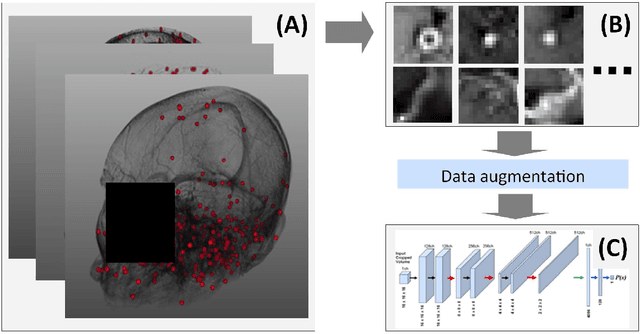
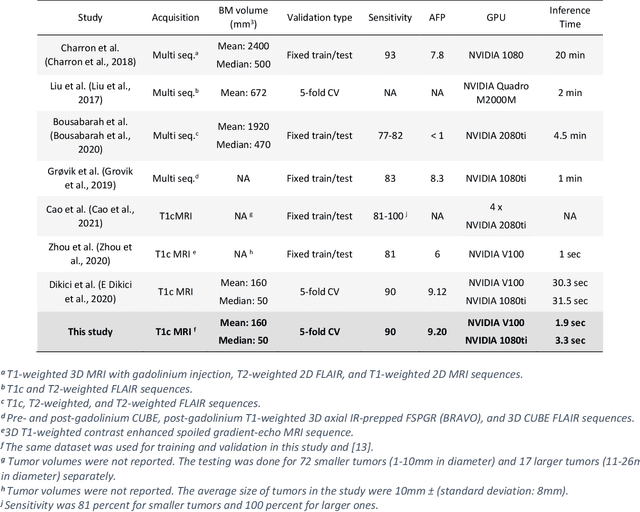
Abstract:Early detection of brain metastases (BM) is one of the determining factors for the successful treatment of patients with cancer; however, the accurate detection of small BM lesions (< 15mm) remains a challenging task. We previously described a framework for the detection of small BM in single-sequence gadolinium-enhanced T1-weighted 3D MRI datasets. It combined classical image processing (IP) with a dedicated convolutional neural network, taking approximately 30 seconds to process each dataset due to computation-intensive IP stages. To overcome the speed limitation, this study aims to reformulate the framework via an augmented pair of CNNs (eliminating the IP) to reduce the processing times while preserving the BM detection performance. Our previous implementation of the BM detection algorithm utilized Laplacian of Gaussians (LoG) for the candidate selection portion of the solution. In this study, we introduce a novel BM candidate detection CNN (cdCNN) to replace this classical IP stage. The network is formulated to have (1) a similar receptive field as the LoG method, and (2) a bias for the detection of BM lesion loci. The proposed CNN is later augmented with a classification CNN to perform the BM detection task. The cdCNN achieved 97.4% BM detection sensitivity when producing 60K candidates per 3D MRI dataset, while the LoG achieved 96.5% detection sensitivity with 73K candidates. The augmented BM detection framework generated on average 9.20 false-positive BM detections per patient for 90% sensitivity, which is comparable with our previous results. However, it processes each 3D data in 1.9 seconds, presenting a 93.5% reduction in the computation time.
Deep Learning-Based Automatic Detection of Poorly Positioned Mammograms to Minimize Patient Return Visits for Repeat Imaging: A Real-World Application
Sep 28, 2020



Abstract:Screening mammograms are a routine imaging exam performed to detect breast cancer in its early stages to reduce morbidity and mortality attributed to this disease. In order to maximize the efficacy of breast cancer screening programs, proper mammographic positioning is paramount. Proper positioning ensures adequate visualization of breast tissue and is necessary for effective breast cancer detection. Therefore, breast-imaging radiologists must assess each mammogram for the adequacy of positioning before providing a final interpretation of the examination; this often necessitates return patient visits for additional imaging. In this paper, we propose a deep learning-algorithm method that mimics and automates this decision-making process to identify poorly positioned mammograms. Our objective for this algorithm is to assist mammography technologists in recognizing inadequately positioned mammograms real-time, improve the quality of mammographic positioning and performance, and ultimately reducing repeat visits for patients with initially inadequate imaging. The proposed model showed a true positive rate for detecting correct positioning of 91.35% in the mediolateral oblique view and 95.11% in the craniocaudal view. In addition to these results, we also present an automatically generated report which can aid the mammography technologist in taking corrective measures during the patient visit.
Artificial Intelligence to Assist in Exclusion of Coronary Atherosclerosis during CCTA Evaluation of Chest-Pain in the Emergency Department: Preparing an Application for Real-World Use
Aug 10, 2020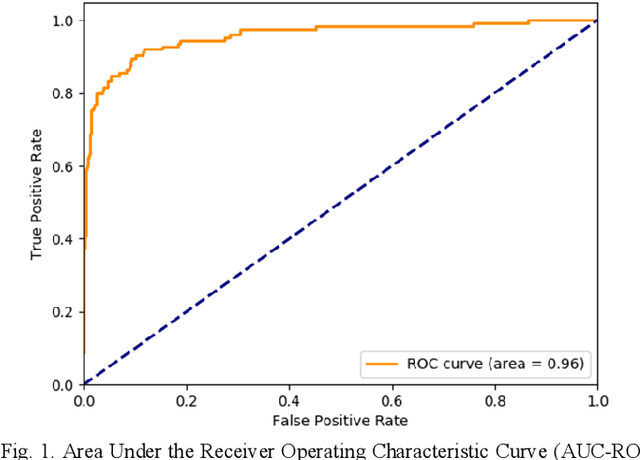
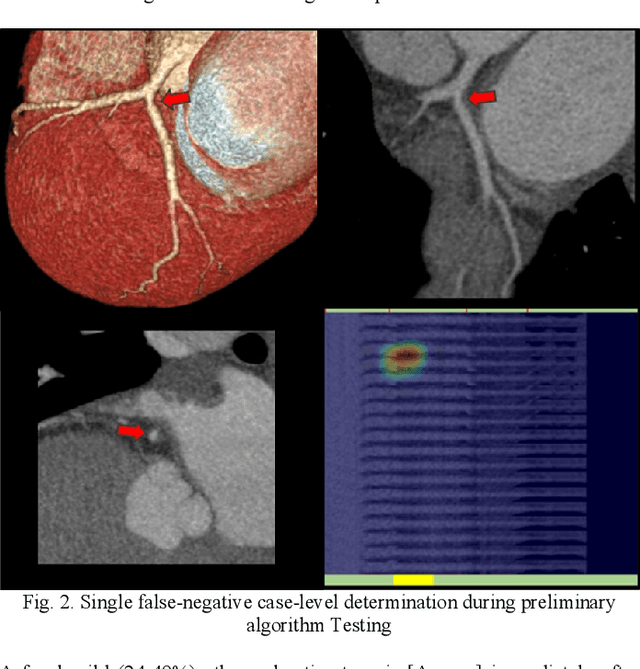
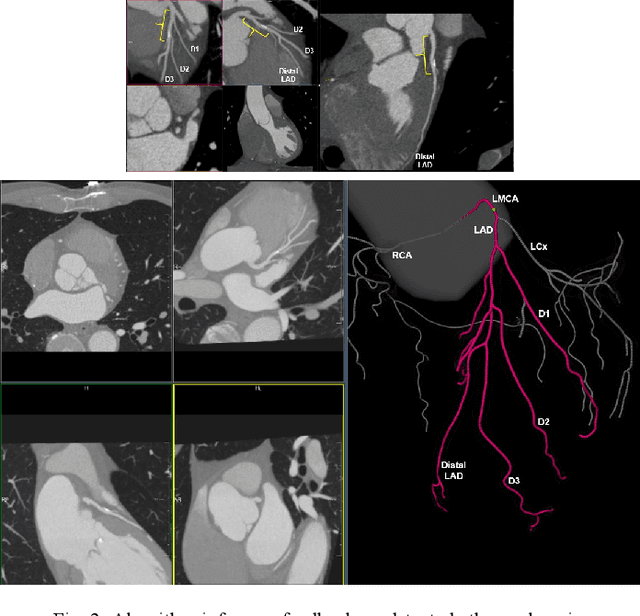
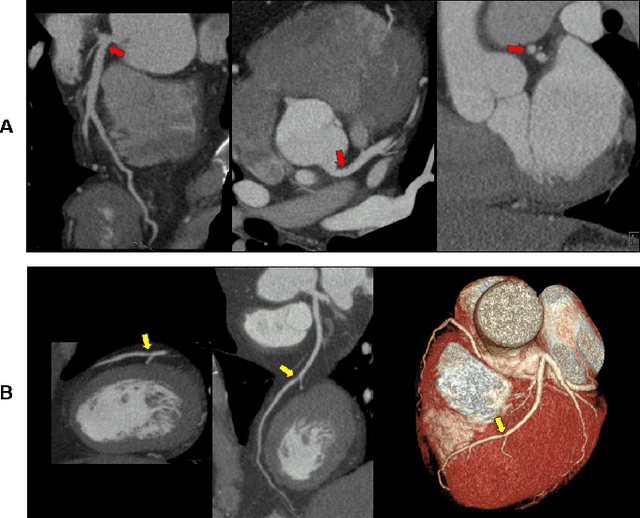
Abstract:Coronary Computed Tomography Angiography (CCTA) evaluation of chest-pain patients in an Emergency Department (ED) is considered appropriate. While a negative CCTA interpretation supports direct patient discharge from an ED, labor-intensive analyses are required, with accuracy in jeopardy from distractions. We describe the development of an Artificial Intelligence (AI) algorithm and workflow for assisting interpreting physicians in CCTA screening for the absence of coronary atherosclerosis. The two-phase approach consisted of (1) Phase 1 - focused on the development and preliminary testing of an algorithm for vessel-centerline extraction classification in a balanced study population (n = 500 with 50% disease prevalence) derived by retrospective random case selection; and (2) Phase 2 - concerned with simulated-clinical Trialing of the developed algorithm on a per-case basis in a more real-world study population (n = 100 with 28% disease prevalence) from an ED chest-pain series. This allowed pre-deployment evaluation of the AI-based CCTA screening application which provides a vessel-by-vessel graphic display of algorithm inference results integrated into a clinically capable viewer. Algorithm performance evaluation used Area Under the Receiver-Operating-Characteristic Curve (AUC-ROC); confusion matrices reflected ground-truth vs AI determinations. The vessel-based algorithm demonstrated strong performance with AUC-ROC = 0.96. In both Phase 1 and Phase 2, independent of disease prevalence differences, negative predictive values at the case level were very high at 95%. The rate of completion of the algorithm workflow process (96% with inference results in 55-80 seconds) in Phase 2 depended on adequate image quality. There is potential for this AI application to assist in CCTA interpretation to help extricate atherosclerosis from chest-pain presentations.
Predicting Rate of Cognitive Decline at Baseline Using a Deep Neural Network with Multidata Analysis
Feb 24, 2020



Abstract:This study investigates whether a machine-learning-based system can predict the rate of cognitive-decline in mildly cognitively impaired (MCI) patients by processing only the clinical and imaging data collected at the initial visit. We build a predictive model based on a supervised hybrid neural network utilizing a 3-Dimensional Convolutional Neural Network to perform volume analysis of Magnetic Resonance Imaging (MRI) and integration of non-imaging clinical data at the fully connected layer of the architecture. The analysis is performed on the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset. Experimental results confirm that there is a correlation between cognitive decline and the data obtained at the first visit. The system achieved an area under the receiver operator curve (AUC) of 66.6% for cognitive decline class prediction.
Are Quantitative Features of Lung Nodules Reproducible at Different CT Acquisition and Reconstruction Parameters?
Aug 14, 2019



Abstract:Consistency and duplicability in Computed Tomography (CT) output is essential to quantitative imaging for lung cancer detection and monitoring. This study of CT-detected lung nodules investigated the reproducibility of volume-, density-, and texture-based features (outcome variables) over routine ranges of radiation-dose, reconstruction kernel, and slice thickness. CT raw data of 23 nodules were reconstructed using 320 acquisition/reconstruction conditions (combinations of 4 doses, 10 kernels, and 8 thicknesses). Scans at 12.5%, 25%, and 50% of protocol dose were simulated; reduced-dose and full-dose data were reconstructed using conventional filtered back-projection and iterative-reconstruction kernels at a range of thicknesses (0.6-5.0 mm). Full-dose/B50f kernel reconstructions underwent expert segmentation for reference Region-Of-Interest (ROI) and nodule volume per thickness; each ROI was applied to 40 corresponding images (combinations of 4 doses and 10 kernels). Typical texture analysis metrics (including 5 histogram features, 13 Gray Level Co-occurrence Matrix, 5 Run Length Matrix, 2 Neighboring Gray-Level Dependence Matrix, and 2 Neighborhood Gray-Tone Difference Matrix) were computed per ROI. Reconstruction conditions resulting in no significant change in volume, density, or texture metrics were identified as "compatible pairs" for a given outcome variable. Our results indicate that as thickness increases, volumetric reproducibility decreases, while reproducibility of histogram- and texture-based features across different acquisition and reconstruction parameters improves. In order to achieve concomitant reproducibility of volumetric and radiomic results across studies, balanced standardization of the imaging acquisition parameters is required.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge