Matthew Bigelow
Real-time CARFAC Cochlea Model Acceleration on FPGA for Underwater Acoustic Sensing Systems
Aug 11, 2025Abstract:This paper presents a real-time, energy-efficient embedded system implementing an array of Cascade of Asymmetric Resonators with Fast-Acting Compression (CARFAC) cochlea models for underwater sound analysis. Built on the AMD Kria KV260 System-on-Module (SoM), the system integrates a Rust-based software framework on the processor for real-time interfacing and synchronization with multiple hydrophone inputs, and a hardware-accelerated implementation of the CARFAC models on a Field-Programmable Gate Array (FPGA) for real-time sound pre-processing. Compared to prior work, the CARFAC accelerator achieves improved scalability and processing speed while reducing resource usage through optimized time-multiplexing, pipelined design, and elimination of costly division circuits. Experimental results demonstrate 13.5% hardware utilization for a single 64-channel CARFAC instance and a whole board power consumption of 3.11 W when processing a 256 kHz input signal in real time.
Advancing Brain Metastases Detection in T1-Weighted Contrast-Enhanced 3D MRI using Noisy Student-based Training
Nov 19, 2021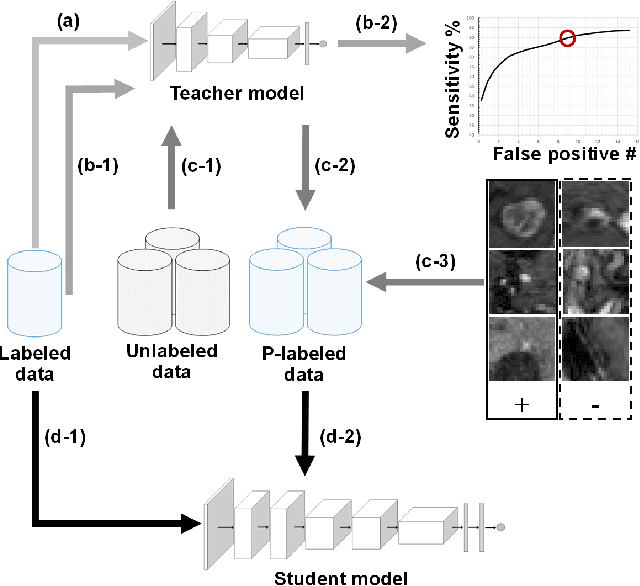
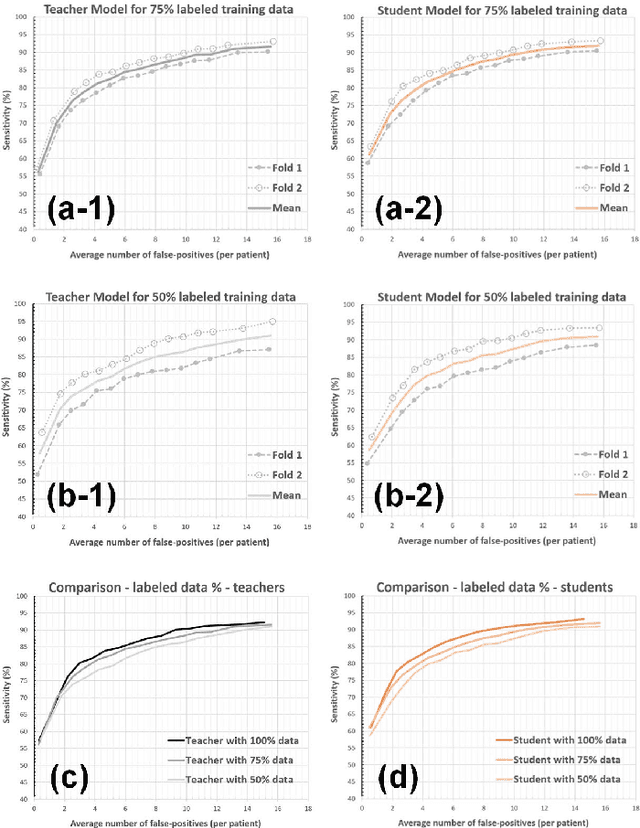
Abstract:The detection of brain metastases (BM) in their early stages could have a positive impact on the outcome of cancer patients. We previously developed a framework for detecting small BM (with diameters of less than 15mm) in T1-weighted Contrast-Enhanced 3D Magnetic Resonance images (T1c) to assist medical experts in this time-sensitive and high-stakes task. The framework utilizes a dedicated convolutional neural network (CNN) trained using labeled T1c data, where the ground truth BM segmentations were provided by a radiologist. This study aims to advance the framework with a noisy student-based self-training strategy to make use of a large corpus of unlabeled T1c data (i.e., data without BM segmentations or detections). Accordingly, the work (1) describes the student and teacher CNN architectures, (2) presents data and model noising mechanisms, and (3) introduces a novel pseudo-labeling strategy factoring in the learned BM detection sensitivity of the framework. Finally, it describes a semi-supervised learning strategy utilizing these components. We performed the validation using 217 labeled and 1247 unlabeled T1c exams via 2-fold cross-validation. The framework utilizing only the labeled exams produced 9.23 false positives for 90% BM detection sensitivity; whereas, the framework using the introduced learning strategy led to ~9% reduction in false detections (i.e., 8.44) for the same sensitivity level. Furthermore, while experiments utilizing 75% and 50% of the labeled datasets resulted in algorithm performance degradation (12.19 and 13.89 false positives respectively), the impact was less pronounced with the noisy student-based training strategy (10.79 and 12.37 false positives respectively).
Augmented Networks for Faster Brain Metastases Detection in T1-Weighted Contrast-Enhanced 3D MRI
May 27, 2021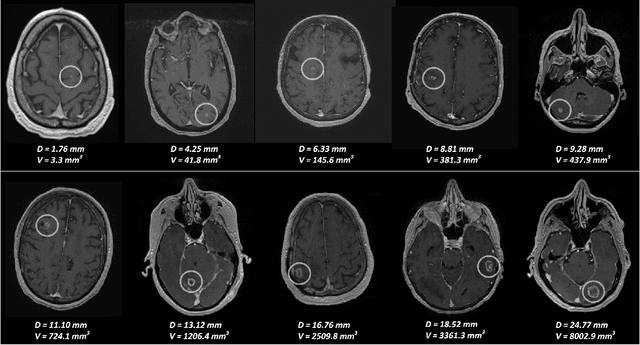
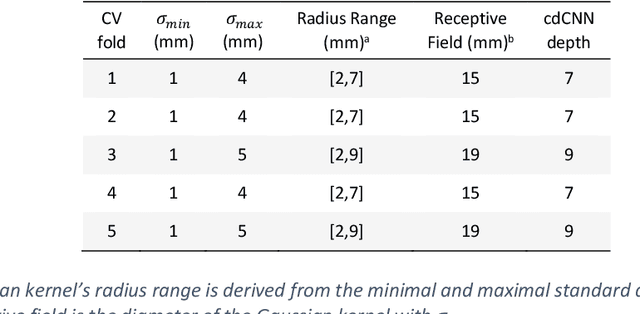
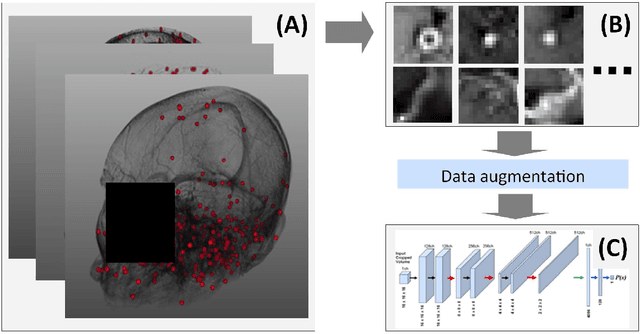
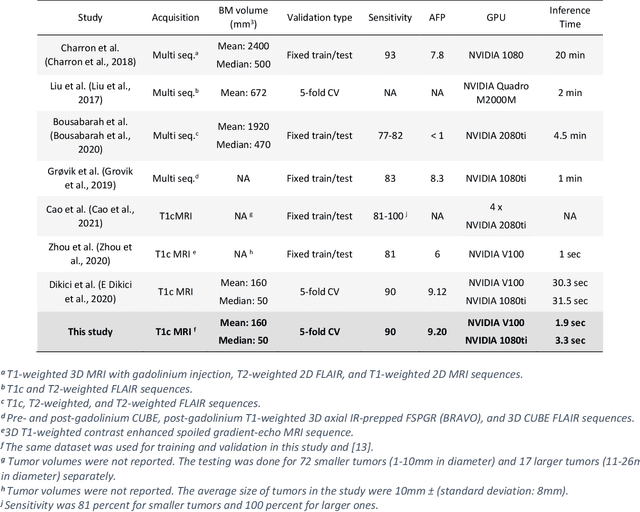
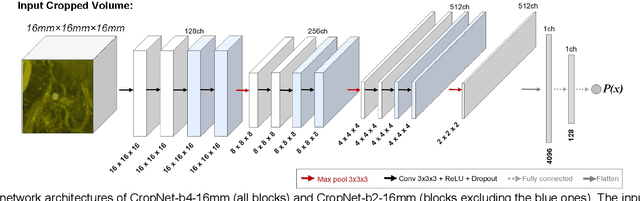
Abstract:Early detection of brain metastases (BM) is one of the determining factors for the successful treatment of patients with cancer; however, the accurate detection of small BM lesions (< 15mm) remains a challenging task. We previously described a framework for the detection of small BM in single-sequence gadolinium-enhanced T1-weighted 3D MRI datasets. It combined classical image processing (IP) with a dedicated convolutional neural network, taking approximately 30 seconds to process each dataset due to computation-intensive IP stages. To overcome the speed limitation, this study aims to reformulate the framework via an augmented pair of CNNs (eliminating the IP) to reduce the processing times while preserving the BM detection performance. Our previous implementation of the BM detection algorithm utilized Laplacian of Gaussians (LoG) for the candidate selection portion of the solution. In this study, we introduce a novel BM candidate detection CNN (cdCNN) to replace this classical IP stage. The network is formulated to have (1) a similar receptive field as the LoG method, and (2) a bias for the detection of BM lesion loci. The proposed CNN is later augmented with a classification CNN to perform the BM detection task. The cdCNN achieved 97.4% BM detection sensitivity when producing 60K candidates per 3D MRI dataset, while the LoG achieved 96.5% detection sensitivity with 73K candidates. The augmented BM detection framework generated on average 9.20 false-positive BM detections per patient for 90% sensitivity, which is comparable with our previous results. However, it processes each 3D data in 1.9 seconds, presenting a 93.5% reduction in the computation time.
Coronary Artery Classification and Weakly Supervised Abnormality Localization on Coronary CT Angiography with 3-Dimensional Convolutional Neural Networks
Nov 26, 2019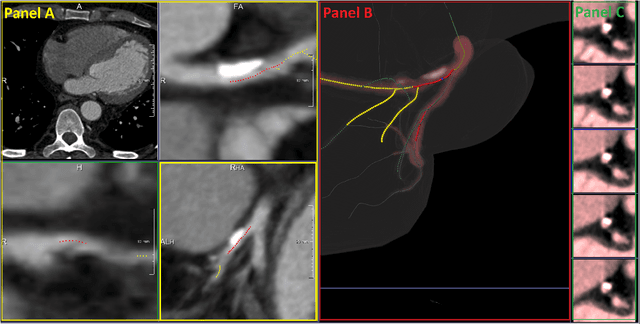
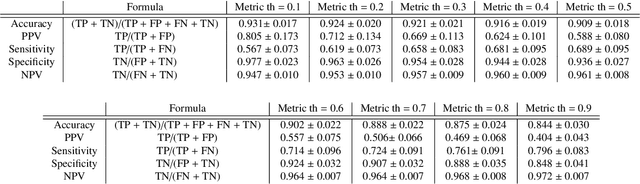
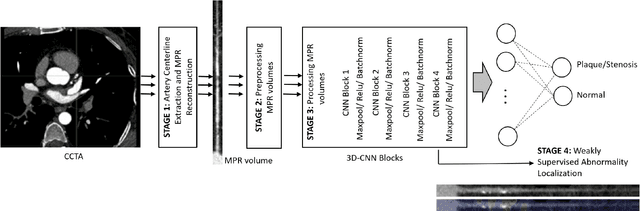
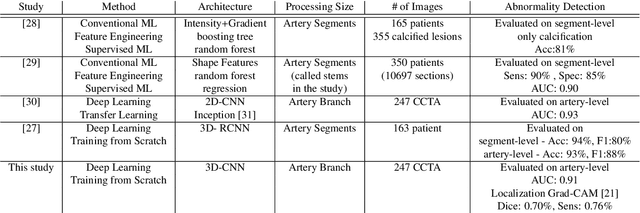
Abstract:We propose a fully automated algorithm based on a deep-learning framework enabling screening of a Coronary Computed Tomography Angiography (CCTA) examination for confident detection of the presence or complete absence of atherosclerotic plaque of the coronary arteries. The system starts with extracting the coronary arteries and their branches from CCTA datasets and representing them with multi-planar reformatted volumes; pre-processing and augmentation techniques are then applied to increase the robustness and generalization ability of the system. A 3-Dimensional Convolutional Neural Network (3D-CNN) is utilized to model pathological changes (e.g., calcification) in coronary arteries/branches. The system then learns the discriminatory features between vessels with and without atherosclerosis. The discriminative features at the final convolutional layer are visualized with a saliency map approach to localize the visual clues related to atherosclerosis. We have evaluated the system on a reference dataset representing 247 patients with atherosclerosis and 246 patients free of atherosclerosis. With 5-fold cross-validation, an accuracy = 90.9%, with Positive Predictive Value = 58.8%, Sensitivity = 68.9%, Specificity of 93.6%, and Negative Predictive Value = 96.1% are achieved at the artery/branch level with a threshold of 0.5. The average area under the curve = 0.91. The system indicates a high negative predictive value, which may be potentially useful for assisting physicians in identifying patients with no coronary atherosclerosis that need no further diagnostic evaluation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge