Yunyan Xing
Multimorbidity Content-Based Medical Image Retrieval Using Proxies
Nov 22, 2022



Abstract:Content-based medical image retrieval is an important diagnostic tool that improves the explainability of computer-aided diagnosis systems and provides decision making support to healthcare professionals. Medical imaging data, such as radiology images, are often multimorbidity; a single sample may have more than one pathology present. As such, image retrieval systems for the medical domain must be designed for the multi-label scenario. In this paper, we propose a novel multi-label metric learning method that can be used for both classification and content-based image retrieval. In this way, our model is able to support diagnosis by predicting the presence of diseases and provide evidence for these predictions by returning samples with similar pathological content to the user. In practice, the retrieved images may also be accompanied by pathology reports, further assisting in the diagnostic process. Our method leverages proxy feature vectors, enabling the efficient learning of a robust feature space in which the distance between feature vectors can be used as a measure of the similarity of those samples. Unlike existing proxy-based methods, training samples are able to assign to multiple proxies that span multiple class labels. This multi-label proxy assignment results in a feature space that encodes the complex relationships between diseases present in medical imaging data. Our method outperforms state-of-the-art image retrieval systems and a set of baseline approaches. We demonstrate the efficacy of our approach to both classification and content-based image retrieval on two multimorbidity radiology datasets.
Adversarial Pulmonary Pathology Translation for Pairwise Chest X-ray Data Augmentation
Oct 11, 2019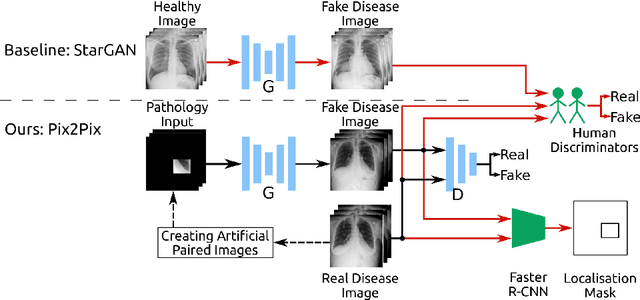
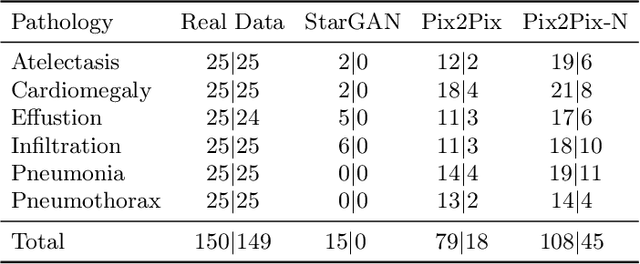
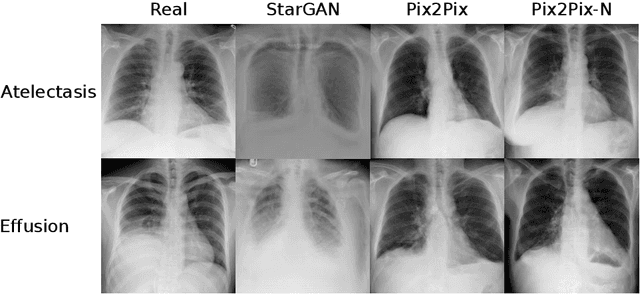

Abstract:Recent works show that Generative Adversarial Networks (GANs) can be successfully applied to chest X-ray data augmentation for lung disease recognition. However, the implausible and distorted pathology features generated from the less than perfect generator may lead to wrong clinical decisions. Why not keep the original pathology region? We proposed a novel approach that allows our generative model to generate high quality plausible images that contain undistorted pathology areas. The main idea is to design a training scheme based on an image-to-image translation network to introduce variations of new lung features around the pathology ground-truth area. Moreover, our model is able to leverage both annotated disease images and unannotated healthy lung images for the purpose of generation. We demonstrate the effectiveness of our model on two tasks: (i) we invite certified radiologists to assess the quality of the generated synthetic images against real and other state-of-the-art generative models, and (ii) data augmentation to improve the performance of disease localisation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge