Jarrel Seah
Adversarial Pulmonary Pathology Translation for Pairwise Chest X-ray Data Augmentation
Oct 11, 2019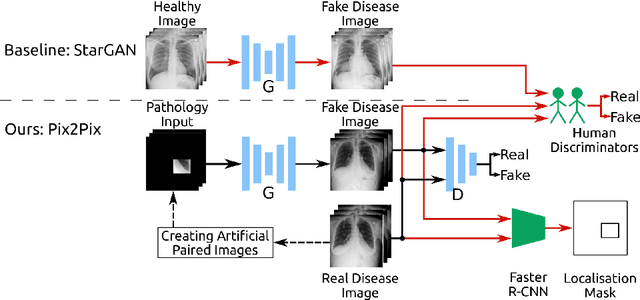
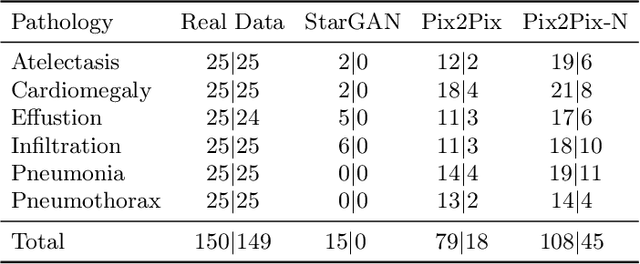
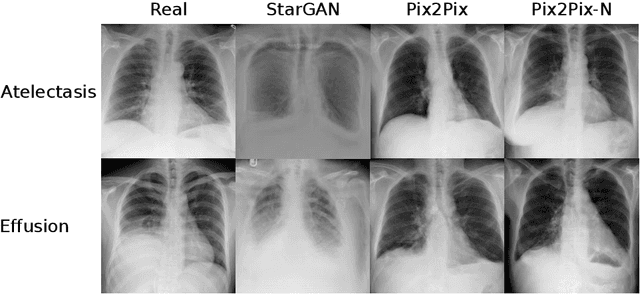

Abstract:Recent works show that Generative Adversarial Networks (GANs) can be successfully applied to chest X-ray data augmentation for lung disease recognition. However, the implausible and distorted pathology features generated from the less than perfect generator may lead to wrong clinical decisions. Why not keep the original pathology region? We proposed a novel approach that allows our generative model to generate high quality plausible images that contain undistorted pathology areas. The main idea is to design a training scheme based on an image-to-image translation network to introduce variations of new lung features around the pathology ground-truth area. Moreover, our model is able to leverage both annotated disease images and unannotated healthy lung images for the purpose of generation. We demonstrate the effectiveness of our model on two tasks: (i) we invite certified radiologists to assess the quality of the generated synthetic images against real and other state-of-the-art generative models, and (ii) data augmentation to improve the performance of disease localisation.
Generative Visual Rationales
Apr 04, 2018
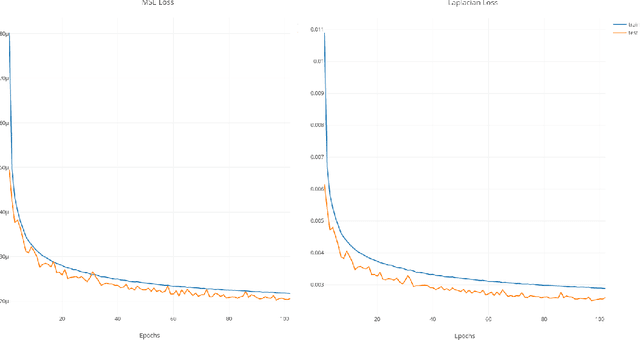
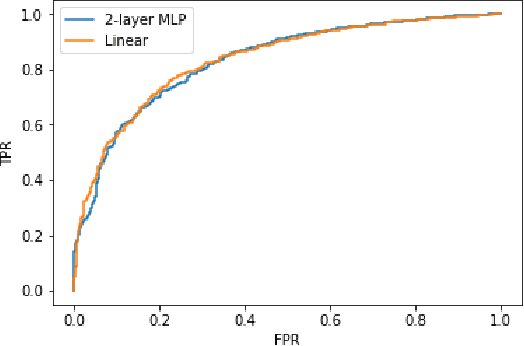
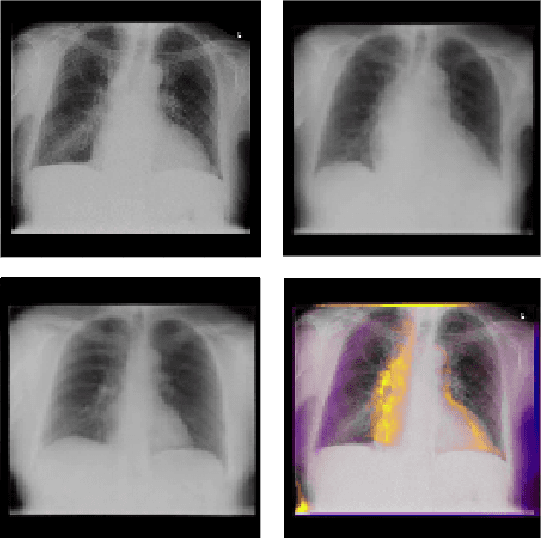
Abstract:Interpretability and small labelled datasets are key issues in the practical application of deep learning, particularly in areas such as medicine. In this paper, we present a semi-supervised technique that addresses both these issues by leveraging large unlabelled datasets to encode and decode images into a dense latent representation. Using chest radiography as an example, we apply this encoder to other labelled datasets and apply simple models to the latent vectors to learn algorithms to identify heart failure. For each prediction, we generate visual rationales by optimizing a latent representation to minimize the prediction of disease while constrained by a similarity measure in image space. Decoding the resultant latent representation produces an image without apparent disease. The difference between the original decoding and the altered image forms an interpretable visual rationale for the algorithm's prediction on that image. We also apply our method to the MNIST dataset and compare the generated rationales to other techniques described in the literature.
Deep Generative Adversarial Neural Networks for Realistic Prostate Lesion MRI Synthesis
Aug 01, 2017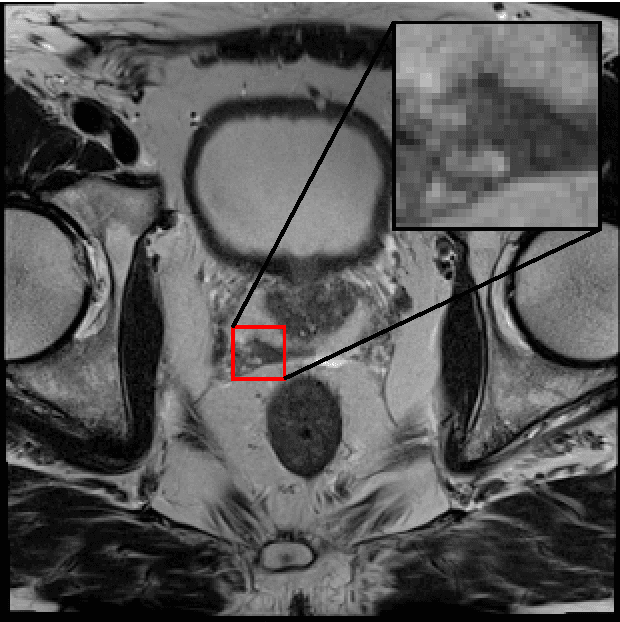
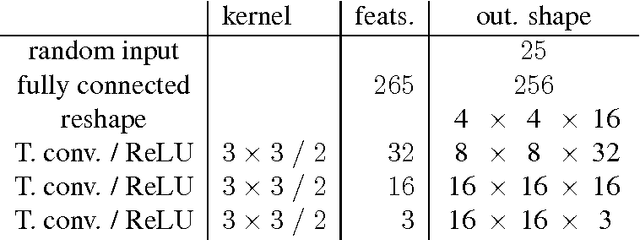
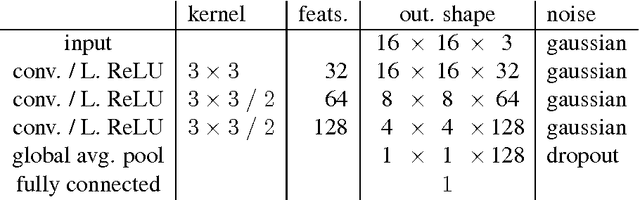
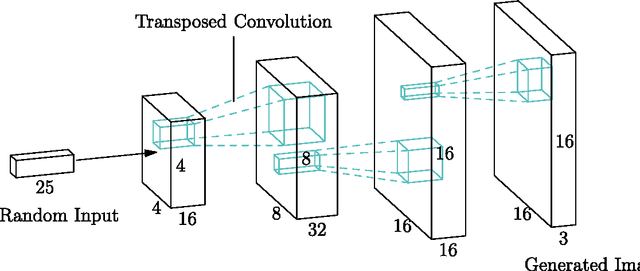
Abstract:Generative Adversarial Neural Networks (GANs) are applied to the synthetic generation of prostate lesion MRI images. GANs have been applied to a variety of natural images, is shown show that the same techniques can be used in the medical domain to create realistic looking synthetic lesion images. 16mm x 16mm patches are extracted from 330 MRI scans from the SPIE ProstateX Challenge 2016 and used to train a Deep Convolutional Generative Adversarial Neural Network (DCGAN) utilizing cutting edge techniques. Synthetic outputs are compared to real images and the implicit latent representations induced by the GAN are explored. Training techniques and successful neural network architectures are explained in detail.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge