Generative Visual Rationales
Paper and Code
Apr 04, 2018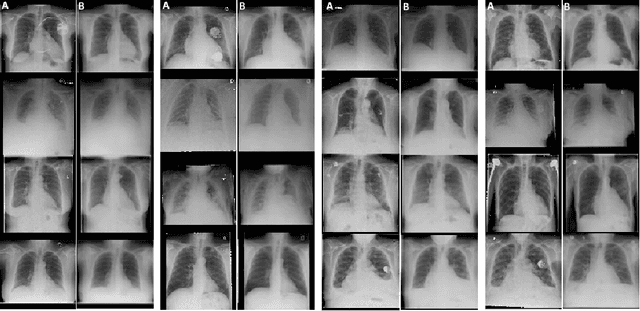
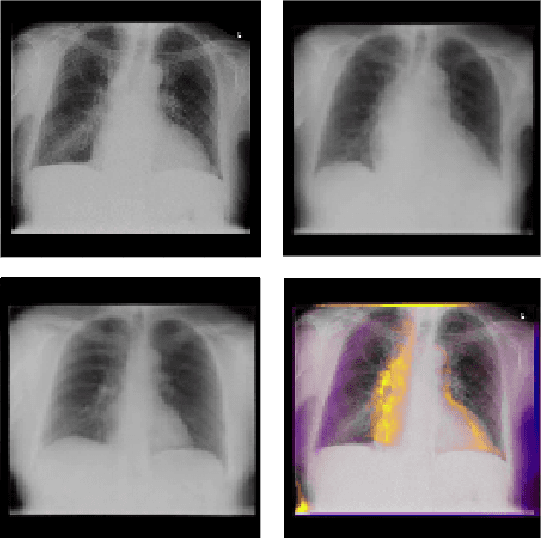
Interpretability and small labelled datasets are key issues in the practical application of deep learning, particularly in areas such as medicine. In this paper, we present a semi-supervised technique that addresses both these issues by leveraging large unlabelled datasets to encode and decode images into a dense latent representation. Using chest radiography as an example, we apply this encoder to other labelled datasets and apply simple models to the latent vectors to learn algorithms to identify heart failure. For each prediction, we generate visual rationales by optimizing a latent representation to minimize the prediction of disease while constrained by a similarity measure in image space. Decoding the resultant latent representation produces an image without apparent disease. The difference between the original decoding and the altered image forms an interpretable visual rationale for the algorithm's prediction on that image. We also apply our method to the MNIST dataset and compare the generated rationales to other techniques described in the literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge