Yongmin Li
Lookahead-then-Verify: Reliable Constrained Decoding for Diffusion LLMs under Context-Free Grammars
Jan 31, 2026Abstract:Diffusion Large Language Models (dLLMs) have demonstrated promising generative capabilities and are increasingly used to produce formal languages defined by context-free grammars, such as source code and chemical expressions. However, as probabilistic models, they still struggle to generate syntactically valid outputs reliably. A natural and promising direction to address this issue is to adapt constrained decoding techniques to enforce grammatical correctness during generation. However, applying these techniques faces two primary obstacles. On the one hand, the non-autoregressive nature of dLLMs renders most existing constrained decoding approaches inapplicable. On the other hand, current approaches specifically designed for dLLMs may allow intermediate outputs that are impossible to complete into valid sentences, which significantly limits their reliability in practice. To address these challenges, we present LAVE, a constrained decoding approach specifically designed for dLLMs. Our approach leverages a key property of dLLMs, namely their ability to predict token distributions for all positions in parallel during each forward pass. Whenever a new token is proposed by model, LAVE performs lookahead using these distributions to efficiently and reliably verify the validity of the proposed token. This design ensures reliable constraints by reliably preserving the potential for intermediate outputs to be extended into valid sentences. Extensive experiments across four widely used dLLMs and three representative benchmarks demonstrate that LAVE consistently outperforms existing baselines and achieves substantial improvements in syntactic correctness, while incurring negligible runtime overhead.
Combining Genre Classification and Harmonic-Percussive Features with Diffusion Models for Music-Video Generation
Dec 07, 2024Abstract:This study presents a novel method for generating music visualisers using diffusion models, combining audio input with user-selected artwork. The process involves two main stages: image generation and video creation. First, music captioning and genre classification are performed, followed by the retrieval of artistic style descriptions. A diffusion model then generates images based on the user's input image and the derived artistic style descriptions. The video generation stage utilises the same diffusion model to interpolate frames, controlled by audio energy vectors derived from key musical features of harmonics and percussives. The method demonstrates promising results across various genres, and a new metric, Audio-Visual Synchrony (AVS), is introduced to quantitatively evaluate the synchronisation between visual and audio elements. Comparative analysis shows significantly higher AVS values for videos generated using the proposed method with audio energy vectors, compared to linear interpolation. This approach has potential applications in diverse fields, including independent music video creation, film production, live music events, and enhancing audio-visual experiences in public spaces.
Showing LLM-Generated Code Selectively Based on Confidence of LLMs
Oct 04, 2024Abstract:Large Language Models (LLMs) have shown impressive abilities in code generation, but they may generate erroneous programs. Reading a program takes ten times longer than writing it. Showing these erroneous programs to developers will waste developers' energies and introduce security risks to software. To address the above limitations, we propose HonestCoder, a novel LLM-based code generation approach. HonestCoder selectively shows the generated programs to developers based on LLMs' confidence. The confidence provides valuable insights into the correctness of generated programs. To achieve this goal, we propose a novel approach to estimate LLMs' confidence in code generation. It estimates confidence by measuring the multi-modal similarity between LLMs-generated programs. We collect and release a multilingual benchmark named TruthCodeBench, which consists of 2,265 samples and covers two popular programming languages (i.e., Python and Java). We apply HonestCoder to four popular LLMs (e.g., DeepSeek-Coder and Code Llama) and evaluate it on TruthCodeBench. Based on the experiments, we obtain the following insights. (1) HonestCoder can effectively estimate LLMs' confidence and accurately determine the correctness of generated programs. For example, HonestCoder outperforms the state-of-the-art baseline by 27.79% in AUROC and 63.74% in AUCPR. (2) HonestCoder can decrease the number of erroneous programs shown to developers. Compared to eight baselines, it can show more correct programs and fewer erroneous programs to developers. (3) Compared to showing code indiscriminately, HonestCoder only adds slight time overhead (approximately 0.4 seconds per requirement). (4) We discuss future directions to facilitate the application of LLMs in software development. We hope this work can motivate broad discussions about measuring the reliability of LLMs' outputs in performing code-related tasks.
Segmenting Medical Images: From UNet to Res-UNet and nnUNet
Jul 05, 2024



Abstract:This study provides a comparative analysis of deep learning models including UNet, Res-UNet, Attention Res-UNet, and nnUNet, and evaluates their performance in brain tumour, polyp, and multi-class heart segmentation tasks. The analysis focuses on precision, accuracy, recall, Dice Similarity Coefficient (DSC), and Intersection over Union (IoU) to assess their clinical applicability. In brain tumour segmentation, Res-UNet and nnUNet significantly outperformed UNet, with Res-UNet leading in DSC and IoU scores, indicating superior accuracy in tumour delineation. Meanwhile, nnUNet excelled in recall and accuracy, which are crucial for reliable tumour detection in clinical diagnosis and planning. In polyp detection, nnUNet was the most effective, achieving the highest metrics across all categories and proving itself as a reliable diagnostic tool in endoscopy. In the complex task of heart segmentation, Res-UNet and Attention Res-UNet were outstanding in delineating the left ventricle, with Res-UNet also leading in right ventricle segmentation. nnUNet was unmatched in myocardium segmentation, achieving top scores in precision, recall, DSC, and IoU. The conclusion notes that although Res-UNet occasionally outperforms nnUNet in specific metrics, the differences are quite small. Moreover, nnUNet consistently shows superior overall performance across the experiments. Particularly noted for its high recall and accuracy, which are crucial in clinical settings to minimize misdiagnosis and ensure timely treatment, nnUNet's robust performance in crucial metrics across all tested categories establishes it as the most effective model for these varied and complex segmentation tasks.
DevEval: A Manually-Annotated Code Generation Benchmark Aligned with Real-World Code Repositories
May 30, 2024Abstract:How to evaluate the coding abilities of Large Language Models (LLMs) remains an open question. We find that existing benchmarks are poorly aligned with real-world code repositories and are insufficient to evaluate the coding abilities of LLMs. To address the knowledge gap, we propose a new benchmark named DevEval, which has three advances. (1) DevEval aligns with real-world repositories in multiple dimensions, e.g., code distributions and dependency distributions. (2) DevEval is annotated by 13 developers and contains comprehensive annotations (e.g., requirements, original repositories, reference code, and reference dependencies). (3) DevEval comprises 1,874 testing samples from 117 repositories, covering 10 popular domains (e.g., Internet, Database). Based on DevEval, we propose repository-level code generation and evaluate 8 popular LLMs on DevEval (e.g., gpt-4, gpt-3.5, StarCoder 2, DeepSeek Coder, CodeLLaMa). Our experiments reveal these LLMs' coding abilities in real-world code repositories. For example, in our experiments, the highest Pass@1 of gpt-4-turbo is only 53.04%. We also analyze LLMs' failed cases and summarize their shortcomings. We hope DevEval can facilitate the development of LLMs in real code repositories. DevEval, prompts, and LLMs' predictions have been released.
Multi-Center Fetal Brain Tissue Annotation (FeTA) Challenge 2022 Results
Feb 08, 2024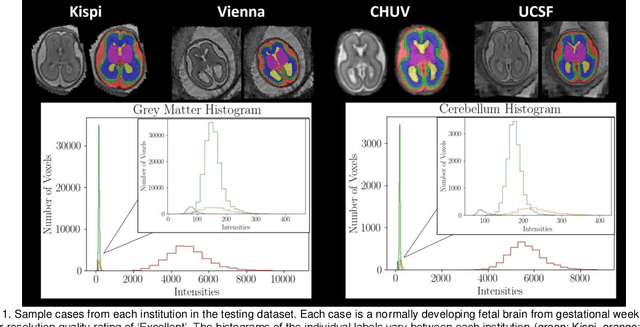
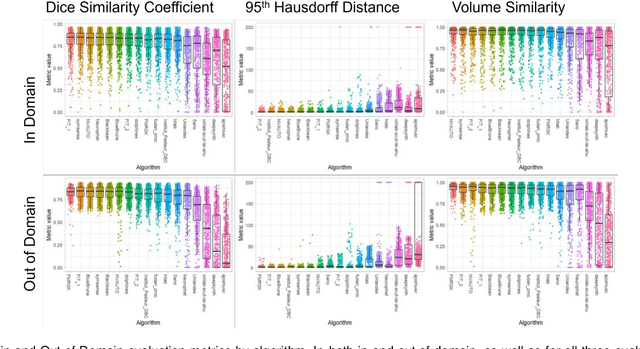
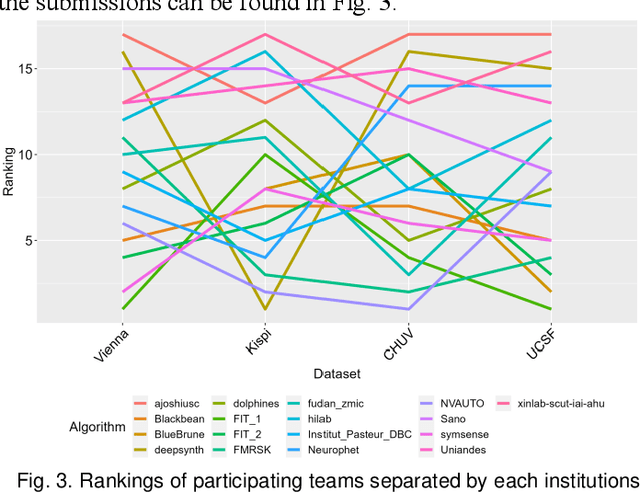
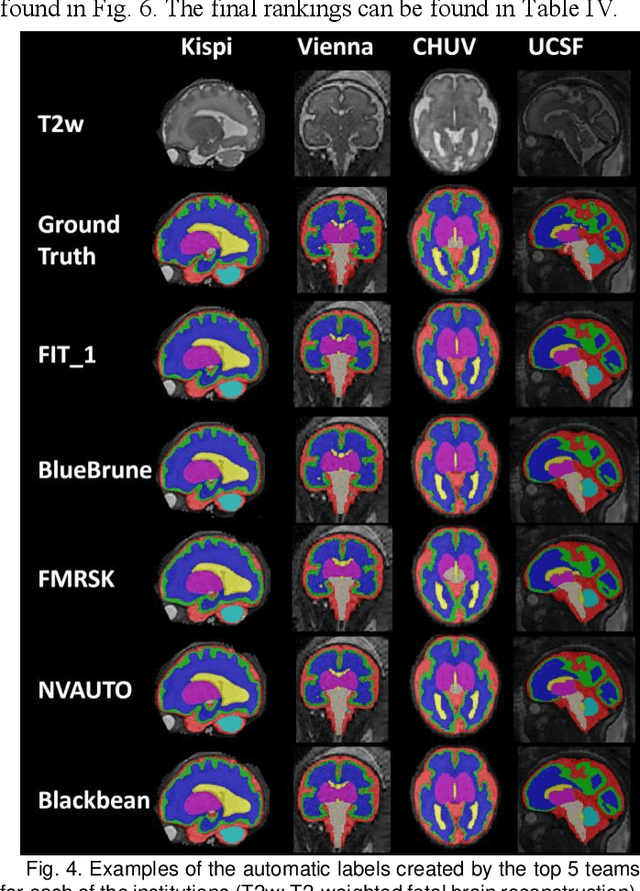
Abstract:Segmentation is a critical step in analyzing the developing human fetal brain. There have been vast improvements in automatic segmentation methods in the past several years, and the Fetal Brain Tissue Annotation (FeTA) Challenge 2021 helped to establish an excellent standard of fetal brain segmentation. However, FeTA 2021 was a single center study, and the generalizability of algorithms across different imaging centers remains unsolved, limiting real-world clinical applicability. The multi-center FeTA Challenge 2022 focuses on advancing the generalizability of fetal brain segmentation algorithms for magnetic resonance imaging (MRI). In FeTA 2022, the training dataset contained images and corresponding manually annotated multi-class labels from two imaging centers, and the testing data contained images from these two imaging centers as well as two additional unseen centers. The data from different centers varied in many aspects, including scanners used, imaging parameters, and fetal brain super-resolution algorithms applied. 16 teams participated in the challenge, and 17 algorithms were evaluated. Here, a detailed overview and analysis of the challenge results are provided, focusing on the generalizability of the submissions. Both in- and out of domain, the white matter and ventricles were segmented with the highest accuracy, while the most challenging structure remains the cerebral cortex due to anatomical complexity. The FeTA Challenge 2022 was able to successfully evaluate and advance generalizability of multi-class fetal brain tissue segmentation algorithms for MRI and it continues to benchmark new algorithms. The resulting new methods contribute to improving the analysis of brain development in utero.
DevEval: Evaluating Code Generation in Practical Software Projects
Jan 26, 2024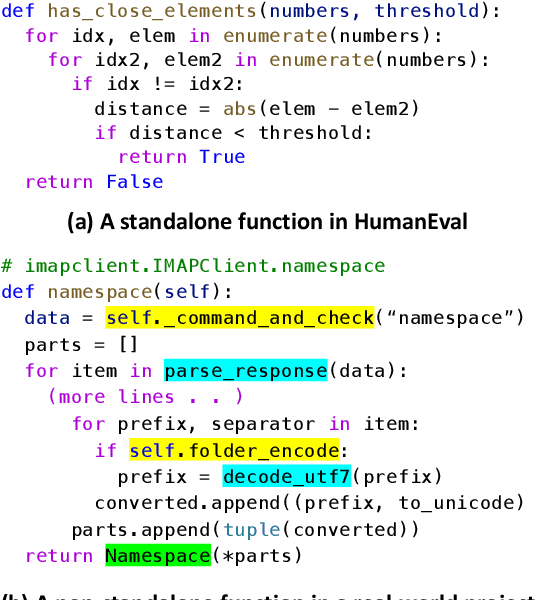

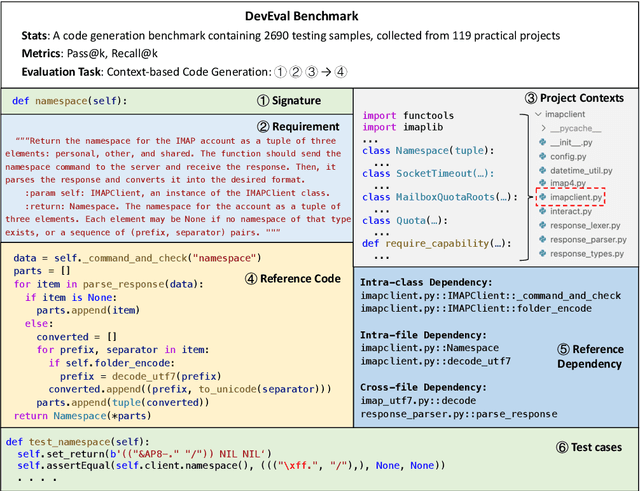
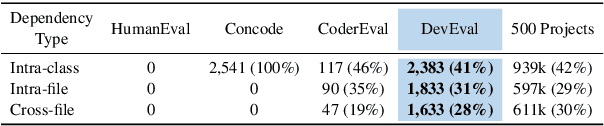
Abstract:How to evaluate Large Language Models (LLMs) in code generation is an open question. Many benchmarks have been proposed but are inconsistent with practical software projects, e.g., unreal program distributions, insufficient dependencies, and small-scale project contexts. Thus, the capabilities of LLMs in practical projects are still unclear. In this paper, we propose a new benchmark named DevEval, aligned with Developers' experiences in practical projects. DevEval is collected through a rigorous pipeline, containing 2,690 samples from 119 practical projects and covering 10 domains. Compared to previous benchmarks, DevEval aligns to practical projects in multiple dimensions, e.g., real program distributions, sufficient dependencies, and enough-scale project contexts. We assess five popular LLMs on DevEval (e.g., gpt-4, gpt-3.5-turbo, CodeLLaMa, and StarCoder) and reveal their actual abilities in code generation. For instance, the highest Pass@1 of gpt-3.5-turbo only is 42 in our experiments. We also discuss the challenges and future directions of code generation in practical projects. We open-source DevEval and hope it can facilitate the development of code generation in practical projects.
SegRap2023: A Benchmark of Organs-at-Risk and Gross Tumor Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Dec 15, 2023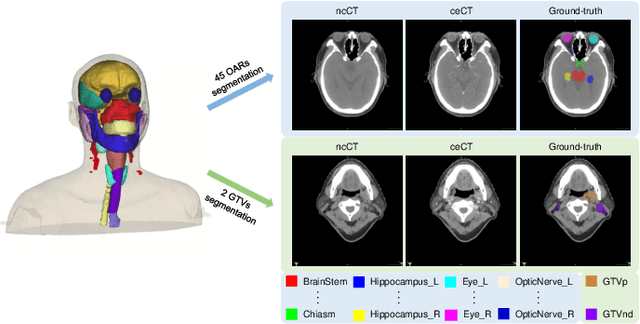
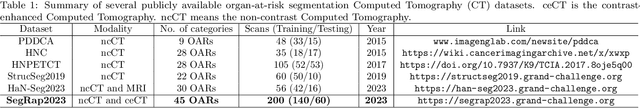
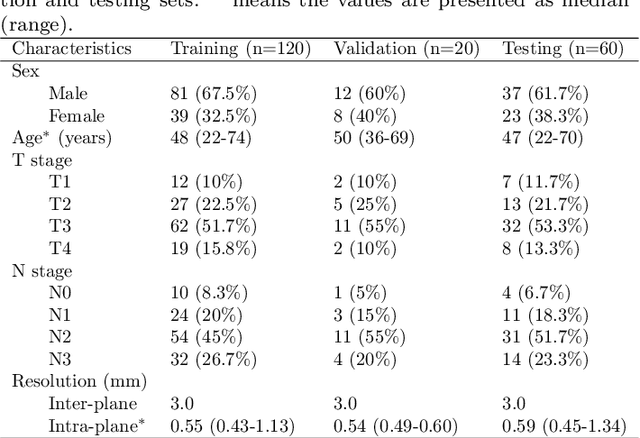
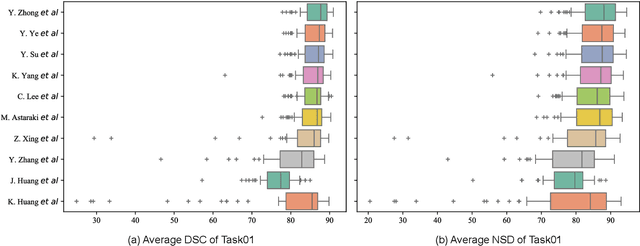
Abstract:Radiation therapy is a primary and effective NasoPharyngeal Carcinoma (NPC) treatment strategy. The precise delineation of Gross Tumor Volumes (GTVs) and Organs-At-Risk (OARs) is crucial in radiation treatment, directly impacting patient prognosis. Previously, the delineation of GTVs and OARs was performed by experienced radiation oncologists. Recently, deep learning has achieved promising results in many medical image segmentation tasks. However, for NPC OARs and GTVs segmentation, few public datasets are available for model development and evaluation. To alleviate this problem, the SegRap2023 challenge was organized in conjunction with MICCAI2023 and presented a large-scale benchmark for OAR and GTV segmentation with 400 Computed Tomography (CT) scans from 200 NPC patients, each with a pair of pre-aligned non-contrast and contrast-enhanced CT scans. The challenge's goal was to segment 45 OARs and 2 GTVs from the paired CT scans. In this paper, we detail the challenge and analyze the solutions of all participants. The average Dice similarity coefficient scores for all submissions ranged from 76.68\% to 86.70\%, and 70.42\% to 73.44\% for OARs and GTVs, respectively. We conclude that the segmentation of large-size OARs is well-addressed, and more efforts are needed for GTVs and small-size or thin-structure OARs. The benchmark will remain publicly available here: https://segrap2023.grand-challenge.org
FashionFlow: Leveraging Diffusion Models for Dynamic Fashion Video Synthesis from Static Imagery
Sep 29, 2023Abstract:Our study introduces a new image-to-video generator called FashionFlow. By utilising a diffusion model, we are able to create short videos from still images. Our approach involves developing and connecting relevant components with the diffusion model, which sets our work apart. The components include the use of pseudo-3D convolutional layers to generate videos efficiently. VAE and CLIP encoders capture vital characteristics from still images to influence the diffusion model. Our research demonstrates a successful synthesis of fashion videos featuring models posing from various angles, showcasing the fit and appearance of the garment. Our findings hold great promise for improving and enhancing the shopping experience for the online fashion industry.
Performance Analysis of UNet and Variants for Medical Image Segmentation
Sep 22, 2023Abstract:Medical imaging plays a crucial role in modern healthcare by providing non-invasive visualisation of internal structures and abnormalities, enabling early disease detection, accurate diagnosis, and treatment planning. This study aims to explore the application of deep learning models, particularly focusing on the UNet architecture and its variants, in medical image segmentation. We seek to evaluate the performance of these models across various challenging medical image segmentation tasks, addressing issues such as image normalization, resizing, architecture choices, loss function design, and hyperparameter tuning. The findings reveal that the standard UNet, when extended with a deep network layer, is a proficient medical image segmentation model, while the Res-UNet and Attention Res-UNet architectures demonstrate smoother convergence and superior performance, particularly when handling fine image details. The study also addresses the challenge of high class imbalance through careful preprocessing and loss function definitions. We anticipate that the results of this study will provide useful insights for researchers seeking to apply these models to new medical imaging problems and offer guidance and best practices for their implementation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge