Alina Miron
Segmenting Medical Images: From UNet to Res-UNet and nnUNet
Jul 05, 2024



Abstract:This study provides a comparative analysis of deep learning models including UNet, Res-UNet, Attention Res-UNet, and nnUNet, and evaluates their performance in brain tumour, polyp, and multi-class heart segmentation tasks. The analysis focuses on precision, accuracy, recall, Dice Similarity Coefficient (DSC), and Intersection over Union (IoU) to assess their clinical applicability. In brain tumour segmentation, Res-UNet and nnUNet significantly outperformed UNet, with Res-UNet leading in DSC and IoU scores, indicating superior accuracy in tumour delineation. Meanwhile, nnUNet excelled in recall and accuracy, which are crucial for reliable tumour detection in clinical diagnosis and planning. In polyp detection, nnUNet was the most effective, achieving the highest metrics across all categories and proving itself as a reliable diagnostic tool in endoscopy. In the complex task of heart segmentation, Res-UNet and Attention Res-UNet were outstanding in delineating the left ventricle, with Res-UNet also leading in right ventricle segmentation. nnUNet was unmatched in myocardium segmentation, achieving top scores in precision, recall, DSC, and IoU. The conclusion notes that although Res-UNet occasionally outperforms nnUNet in specific metrics, the differences are quite small. Moreover, nnUNet consistently shows superior overall performance across the experiments. Particularly noted for its high recall and accuracy, which are crucial in clinical settings to minimize misdiagnosis and ensure timely treatment, nnUNet's robust performance in crucial metrics across all tested categories establishes it as the most effective model for these varied and complex segmentation tasks.
Multi-Center Fetal Brain Tissue Annotation (FeTA) Challenge 2022 Results
Feb 08, 2024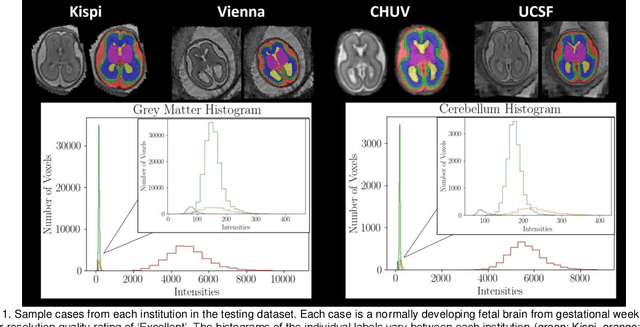
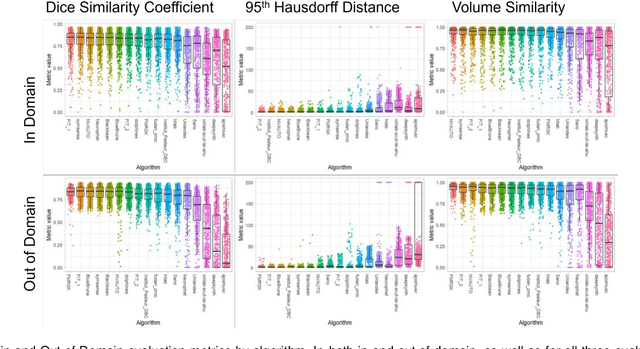
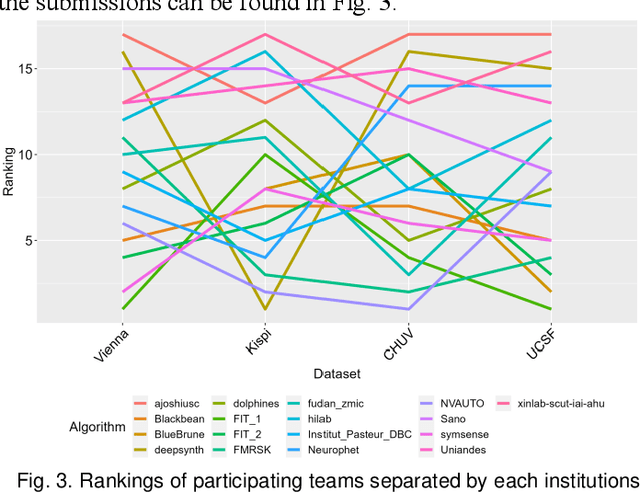
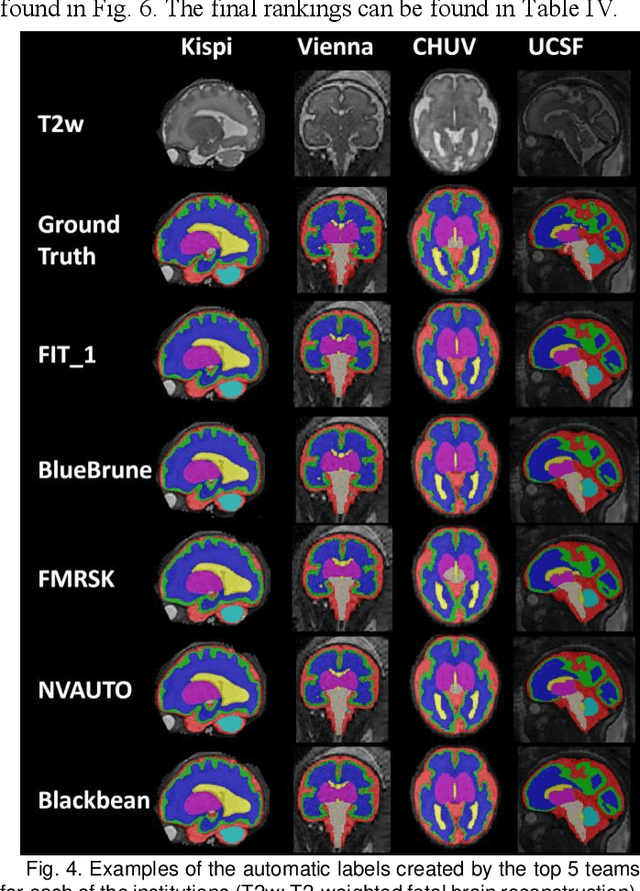
Abstract:Segmentation is a critical step in analyzing the developing human fetal brain. There have been vast improvements in automatic segmentation methods in the past several years, and the Fetal Brain Tissue Annotation (FeTA) Challenge 2021 helped to establish an excellent standard of fetal brain segmentation. However, FeTA 2021 was a single center study, and the generalizability of algorithms across different imaging centers remains unsolved, limiting real-world clinical applicability. The multi-center FeTA Challenge 2022 focuses on advancing the generalizability of fetal brain segmentation algorithms for magnetic resonance imaging (MRI). In FeTA 2022, the training dataset contained images and corresponding manually annotated multi-class labels from two imaging centers, and the testing data contained images from these two imaging centers as well as two additional unseen centers. The data from different centers varied in many aspects, including scanners used, imaging parameters, and fetal brain super-resolution algorithms applied. 16 teams participated in the challenge, and 17 algorithms were evaluated. Here, a detailed overview and analysis of the challenge results are provided, focusing on the generalizability of the submissions. Both in- and out of domain, the white matter and ventricles were segmented with the highest accuracy, while the most challenging structure remains the cerebral cortex due to anatomical complexity. The FeTA Challenge 2022 was able to successfully evaluate and advance generalizability of multi-class fetal brain tissue segmentation algorithms for MRI and it continues to benchmark new algorithms. The resulting new methods contribute to improving the analysis of brain development in utero.
SegRap2023: A Benchmark of Organs-at-Risk and Gross Tumor Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Dec 15, 2023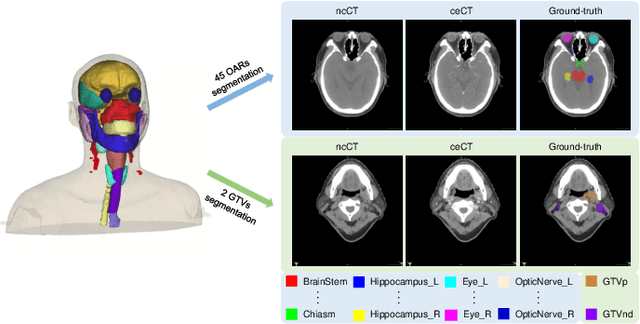
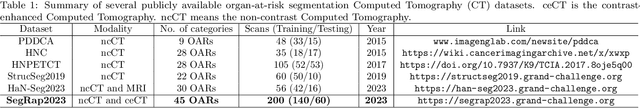
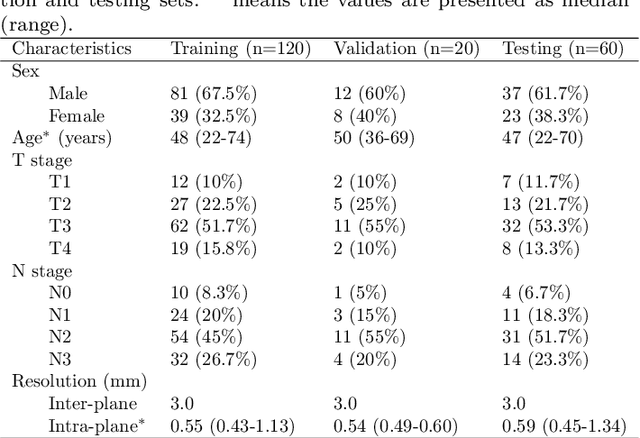
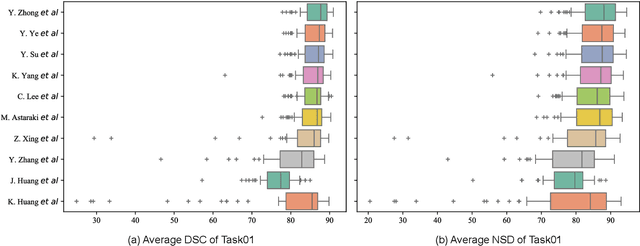
Abstract:Radiation therapy is a primary and effective NasoPharyngeal Carcinoma (NPC) treatment strategy. The precise delineation of Gross Tumor Volumes (GTVs) and Organs-At-Risk (OARs) is crucial in radiation treatment, directly impacting patient prognosis. Previously, the delineation of GTVs and OARs was performed by experienced radiation oncologists. Recently, deep learning has achieved promising results in many medical image segmentation tasks. However, for NPC OARs and GTVs segmentation, few public datasets are available for model development and evaluation. To alleviate this problem, the SegRap2023 challenge was organized in conjunction with MICCAI2023 and presented a large-scale benchmark for OAR and GTV segmentation with 400 Computed Tomography (CT) scans from 200 NPC patients, each with a pair of pre-aligned non-contrast and contrast-enhanced CT scans. The challenge's goal was to segment 45 OARs and 2 GTVs from the paired CT scans. In this paper, we detail the challenge and analyze the solutions of all participants. The average Dice similarity coefficient scores for all submissions ranged from 76.68\% to 86.70\%, and 70.42\% to 73.44\% for OARs and GTVs, respectively. We conclude that the segmentation of large-size OARs is well-addressed, and more efforts are needed for GTVs and small-size or thin-structure OARs. The benchmark will remain publicly available here: https://segrap2023.grand-challenge.org
FashionFlow: Leveraging Diffusion Models for Dynamic Fashion Video Synthesis from Static Imagery
Sep 29, 2023Abstract:Our study introduces a new image-to-video generator called FashionFlow. By utilising a diffusion model, we are able to create short videos from still images. Our approach involves developing and connecting relevant components with the diffusion model, which sets our work apart. The components include the use of pseudo-3D convolutional layers to generate videos efficiently. VAE and CLIP encoders capture vital characteristics from still images to influence the diffusion model. Our research demonstrates a successful synthesis of fashion videos featuring models posing from various angles, showcasing the fit and appearance of the garment. Our findings hold great promise for improving and enhancing the shopping experience for the online fashion industry.
Retinal Image Segmentation with Small Datasets
Mar 09, 2023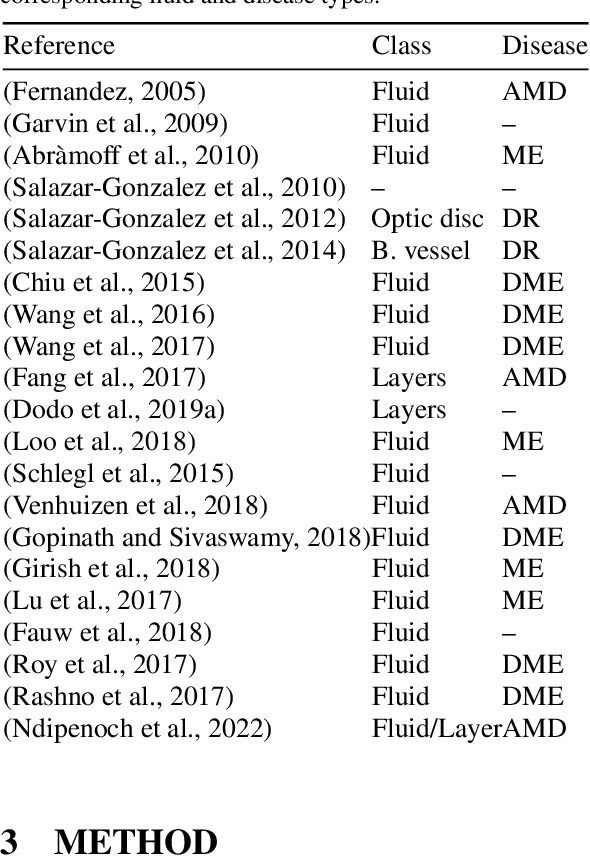
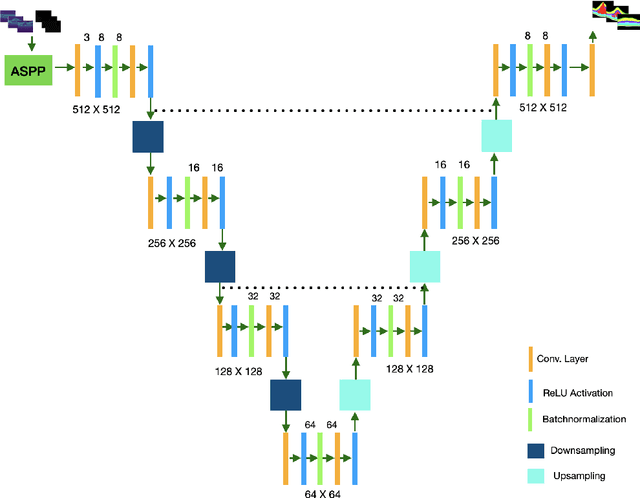
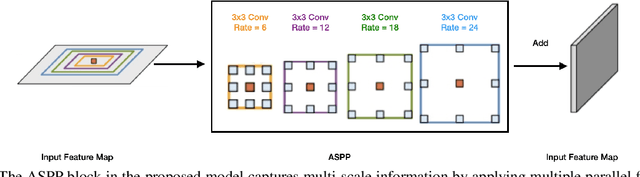
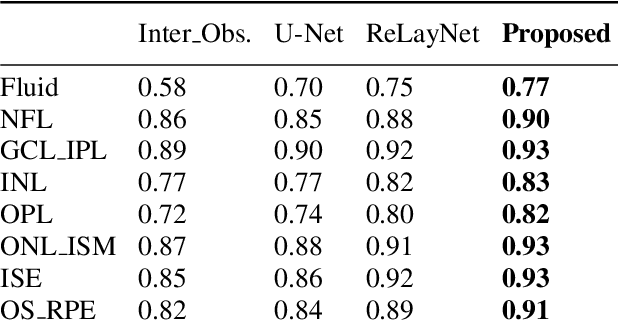
Abstract:Many eye diseases like Diabetic Macular Edema (DME), Age-related Macular Degeneration (AMD), and Glaucoma manifest in the retina, can cause irreversible blindness or severely impair the central version. The Optical Coherence Tomography (OCT), a 3D scan of the retina with high qualitative information about the retinal morphology, can be used to diagnose and monitor changes in the retinal anatomy. Many Deep Learning (DL) methods have shared the success of developing an automated tool to monitor pathological changes in the retina. However, the success of these methods depend mainly on large datasets. To address the challenge from very small and limited datasets, we proposed a DL architecture termed CoNet (Coherent Network) for joint segmentation of layers and fluids in retinal OCT images on very small datasets (less than a hundred training samples). The proposed model was evaluated on the publicly available Duke DME dataset consisting of 110 B-Scans from 10 patients suffering from DME. Experimental results show that the proposed model outperformed both the human experts' annotation and the current state-of-the-art architectures by a clear margin with a mean Dice Score of 88% when trained on 55 images without any data augmentation.
nnUNet RASPP for Retinal OCT Fluid Detection, Segmentation and Generalisation over Variations of Data Sources
Feb 25, 2023Abstract:Retinal Optical Coherence Tomography (OCT), a noninvasive cross-sectional scan of the eye with qualitative 3D visualization of the retinal anatomy is use to study the retinal structure and the presence of pathogens. The advent of the retinal OCT has transformed ophthalmology and it is currently paramount for the diagnosis, monitoring and treatment of many eye pathogens including Macular Edema which impairs vision severely or Glaucoma that can cause irreversible blindness. However the quality of retinal OCT images varies among device manufacturers. Deep Learning methods have had their success in the medical image segmentation community but it is still not clear if the level of success can be generalised across OCT images collected from different device vendors. In this work we propose two variants of the nnUNet [8]. The standard nnUNet and an enhanced vision call nnUnet_RASPP (nnU-Net with residual and Atrous Spatial Pyramid Pooling) both of which are robust and generalise with consistent high performance across images from multiple device vendors. The algorithm was validated on the MICCAI 2017 RETOUCH challenge dataset [1] acquired from 3 device vendors across 3 medical centers from patients suffering from 2 retinal disease types. Experimental results show that our algorithms outperform the current state-of-the-arts algorithms by a clear margin for segmentation obtaining a mean Dice Score (DS) of 82.3% for the 3 retinal fluids scoring 84.0%, 80.0%, 83.0% for Intraretinal Fluid (IRF), Subretinal Fluid (SRF), and Pigment Epithelium Detachments (PED) respectively on the testing dataset. Also we obtained a perfect Area Under the Curve (AUC) score of 100% for the detection of the presence of fluid for all 3 fluid classes on the testing dataset.
Classifying action correctness in physical rehabilitation exercises
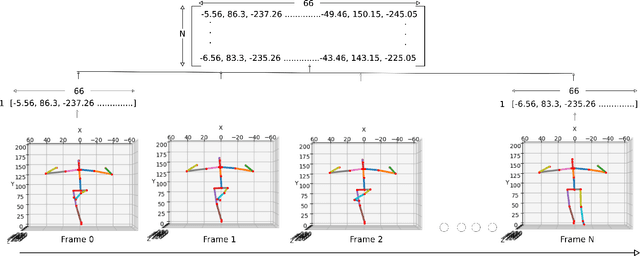
Aug 03, 2021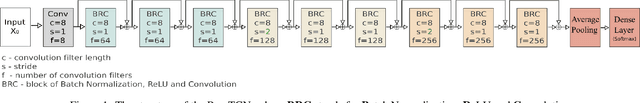
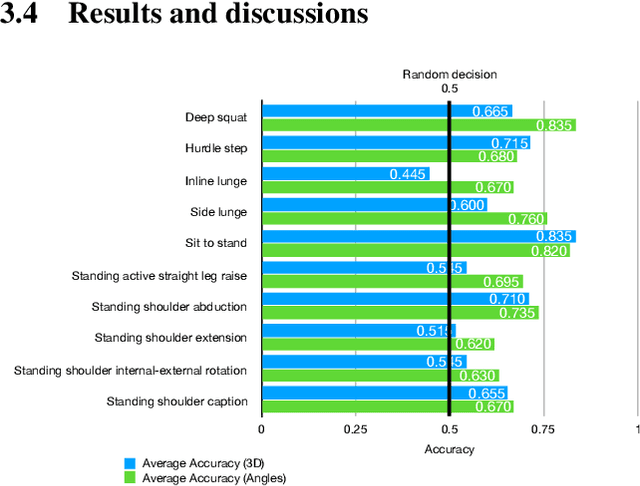
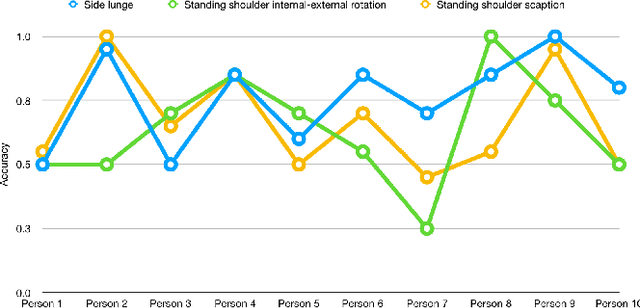
Abstract:The work in this paper focuses on the role of machine learning in assessing the correctness of a human motion or action. This task proves to be more challenging than the gesture and action recognition ones. We will demonstrate, through a set of experiments on a recent dataset, that machine learning algorithms can produce good results for certain actions, but can also fall into the trap of classifying an incorrect execution of an action as a correct execution of another action.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge