Yong Zhu
Pangu Ultra MoE: How to Train Your Big MoE on Ascend NPUs
May 07, 2025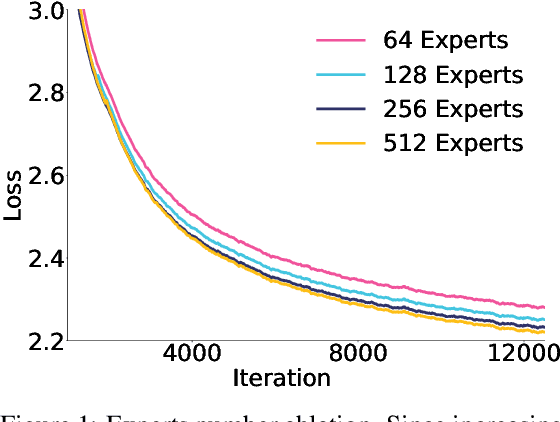

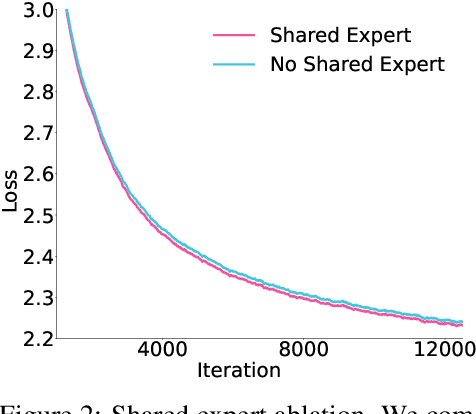

Abstract:Sparse large language models (LLMs) with Mixture of Experts (MoE) and close to a trillion parameters are dominating the realm of most capable language models. However, the massive model scale poses significant challenges for the underlying software and hardware systems. In this paper, we aim to uncover a recipe to harness such scale on Ascend NPUs. The key goals are better usage of the computing resources under the dynamic sparse model structures and materializing the expected performance gain on the actual hardware. To select model configurations suitable for Ascend NPUs without repeatedly running the expensive experiments, we leverage simulation to compare the trade-off of various model hyperparameters. This study led to Pangu Ultra MoE, a sparse LLM with 718 billion parameters, and we conducted experiments on the model to verify the simulation results. On the system side, we dig into Expert Parallelism to optimize the communication between NPU devices to reduce the synchronization overhead. We also optimize the memory efficiency within the devices to further reduce the parameter and activation management overhead. In the end, we achieve an MFU of 30.0% when training Pangu Ultra MoE, with performance comparable to that of DeepSeek R1, on 6K Ascend NPUs, and demonstrate that the Ascend system is capable of harnessing all the training stages of the state-of-the-art language models. Extensive experiments indicate that our recipe can lead to efficient training of large-scale sparse language models with MoE. We also study the behaviors of such models for future reference.
SDSAT: Accelerating LLM Inference through Speculative Decoding with Semantic Adaptive Tokens
Apr 01, 2024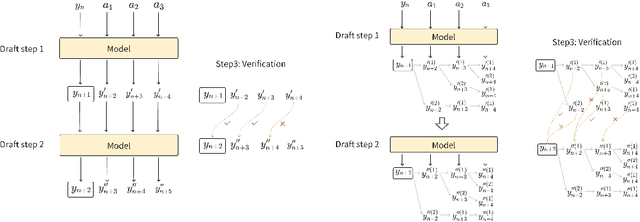
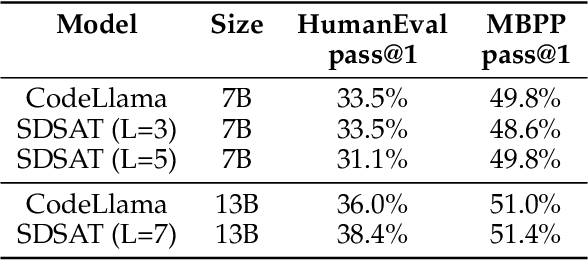
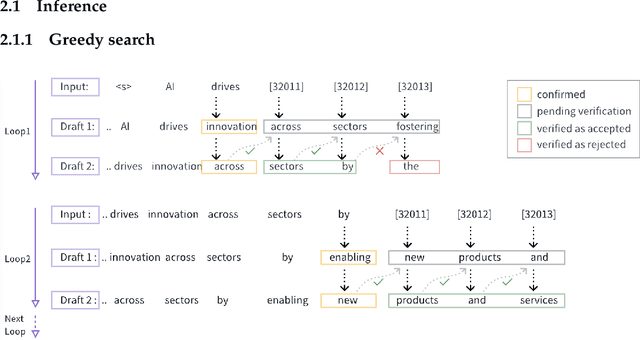
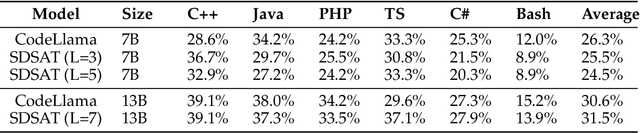
Abstract:We propose an acceleration scheme for large language models (LLMs) through Speculative Decoding with Semantic Adaptive Tokens (SDSAT). The primary objective of this design is to enhance the LLM model's ability to generate draft tokens more accurately without compromising the model's accuracy. The core strategies involve: 1) Fine-tune the model by incorporating semantic adaptive tokens that possess flexible decoding capabilities without changing its structure, allowing them to generate high-quality draft tokens. 2) By employing a training method that does not affect the standard tokens, the model can acquire parallel decoding abilities atop its original framework with minimal training overhead. 3) We have designed the "two-step-draft-then-verify" generation strategies using both greedy search and nucleus sampling. Experiments conducted on the CodeLlama-13B and 7B models have yielded speed increases of over 3.5X and 3.0X, respectively. Please refer to https://github.com/hasuoshenyun/SDSAT.
Mixture of Experts for Biomedical Question Answering
Apr 15, 2022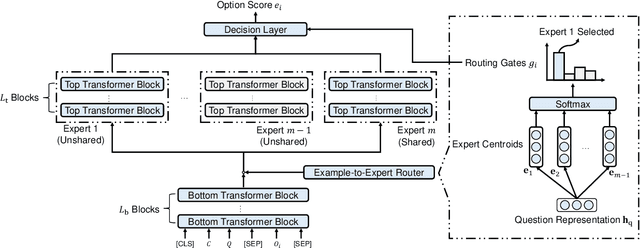
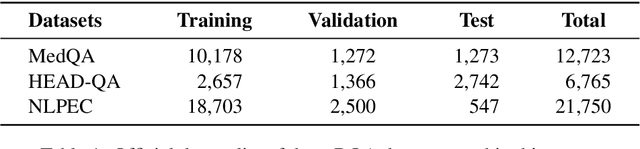

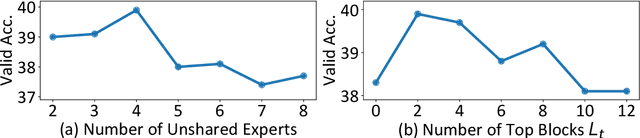
Abstract:Biomedical Question Answering (BQA) has attracted increasing attention in recent years due to its promising application prospect. It is a challenging task because the biomedical questions are professional and usually vary widely. Existing question answering methods answer all questions with a homogeneous model, leading to various types of questions competing for the shared parameters, which will confuse the model decision for each single type of questions. In this paper, in order to alleviate the parameter competition problem, we propose a Mixture-of-Expert (MoE) based question answering method called MoEBQA that decouples the computation for different types of questions by sparse routing. To be specific, we split a pretrained Transformer model into bottom and top blocks. The bottom blocks are shared by all the examples, aiming to capture the general features. The top blocks are extended to an MoE version that consists of a series of independent experts, where each example is assigned to a few experts according to its underlying question type. MoEBQA automatically learns the routing strategy in an end-to-end manner so that each expert tends to deal with the question types it is expert in. We evaluate MoEBQA on three BQA datasets constructed based on real examinations. The results show that our MoE extension significantly boosts the performance of question answering models and achieves new state-of-the-art performance. In addition, we elaborately analyze our MoE modules to reveal how MoEBQA works and find that it can automatically group the questions into human-readable clusters.
Complex Evolutional Pattern Learning for Temporal Knowledge Graph Reasoning
Mar 20, 2022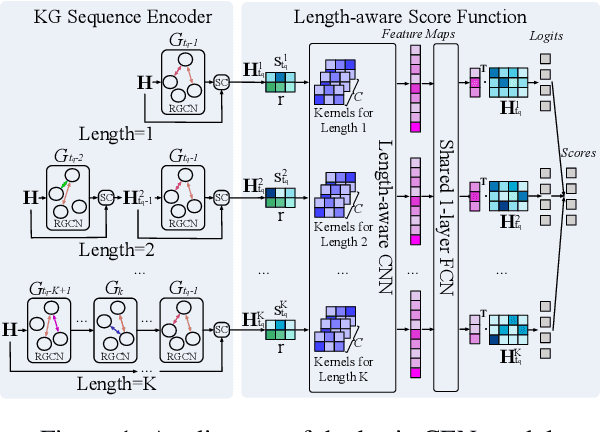
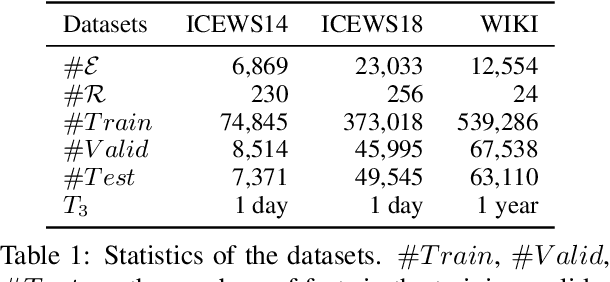
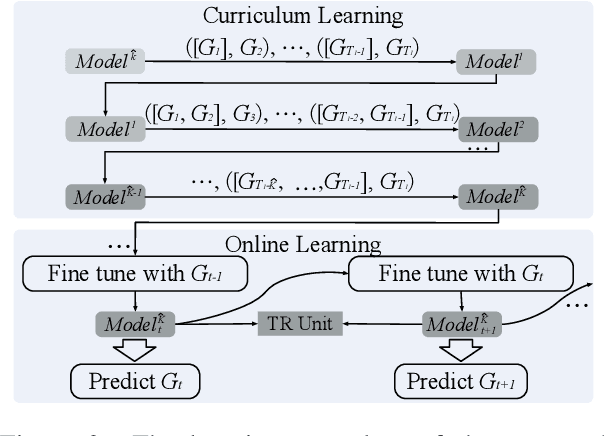
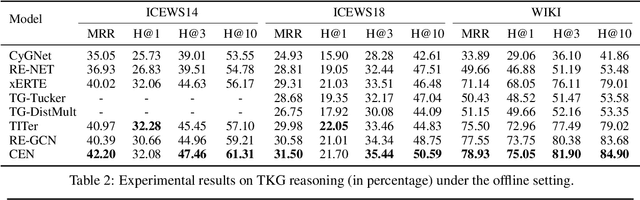
Abstract:A Temporal Knowledge Graph (TKG) is a sequence of KGs corresponding to different timestamps. TKG reasoning aims to predict potential facts in the future given the historical KG sequences. One key of this task is to mine and understand evolutional patterns of facts from these sequences. The evolutional patterns are complex in two aspects, length-diversity and time-variability. Existing models for TKG reasoning focus on modeling fact sequences of a fixed length, which cannot discover complex evolutional patterns that vary in length. Furthermore, these models are all trained offline, which cannot well adapt to the changes of evolutional patterns from then on. Thus, we propose a new model, called Complex Evolutional Network (CEN), which uses a length-aware Convolutional Neural Network (CNN) to handle evolutional patterns of different lengths via an easy-to-difficult curriculum learning strategy. Besides, we propose to learn the model under the online setting so that it can adapt to the changes of evolutional patterns over time. Extensive experiments demonstrate that CEN obtains substantial performance improvement under both the traditional offline and the proposed online settings.
Building Chinese Biomedical Language Models via Multi-Level Text Discrimination
Oct 14, 2021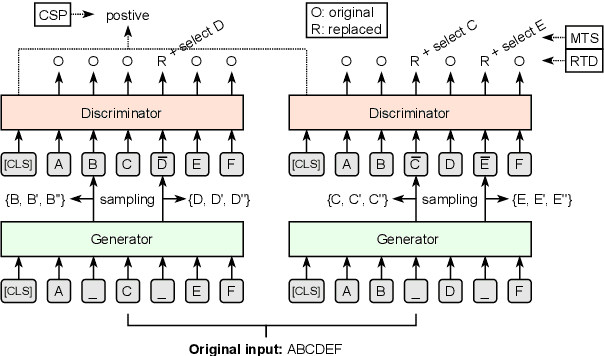
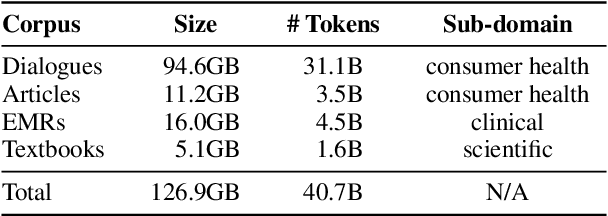
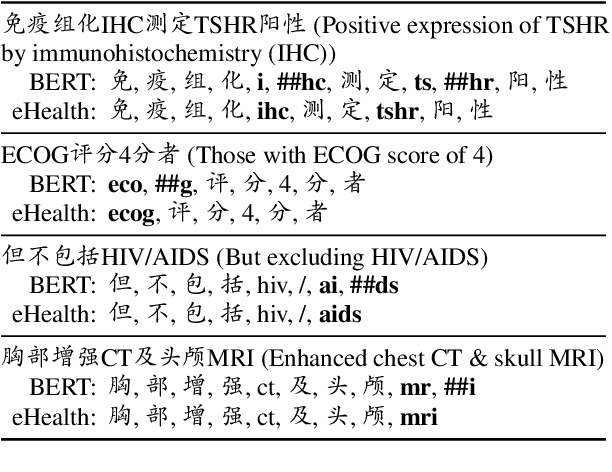
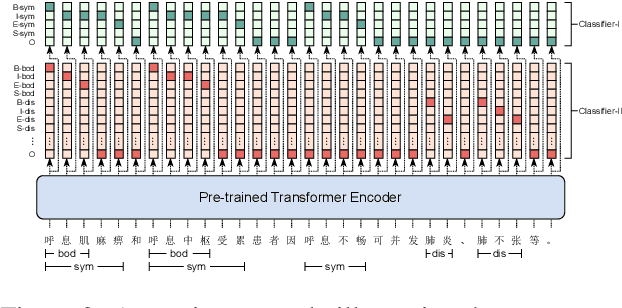
Abstract:Pre-trained language models (PLMs), such as BERT and GPT, have revolutionized the field of NLP, not only in the general domain but also in the biomedical domain. Most prior efforts in building biomedical PLMs have resorted simply to domain adaptation and focused mainly on English. In this work we introduce eHealth, a biomedical PLM in Chinese built with a new pre-training framework. This new framework trains eHealth as a discriminator through both token-level and sequence-level discrimination. The former is to detect input tokens corrupted by a generator and select their original signals from plausible candidates, while the latter is to further distinguish corruptions of a same original sequence from those of the others. As such, eHealth can learn language semantics at both the token and sequence levels. Extensive experiments on 11 Chinese biomedical language understanding tasks of various forms verify the effectiveness and superiority of our approach. The pre-trained model is available to the public at \url{https://github.com/PaddlePaddle/Research/tree/master/KG/eHealth} and the code will also be released later.
Link Prediction on N-ary Relational Facts: A Graph-based Approach
May 18, 2021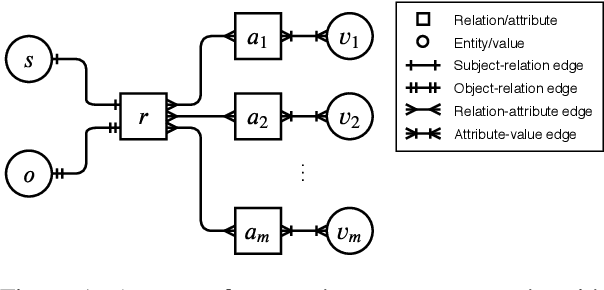
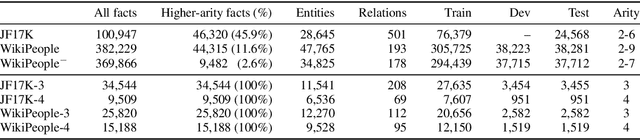
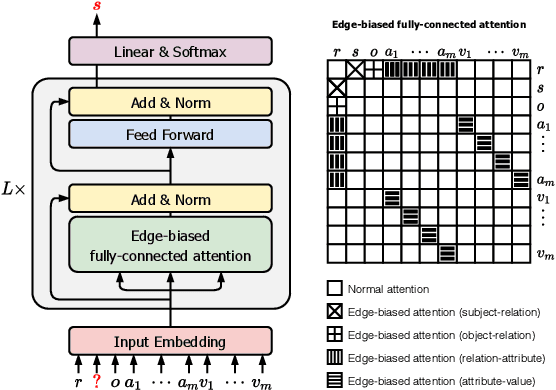

Abstract:Link prediction on knowledge graphs (KGs) is a key research topic. Previous work mainly focused on binary relations, paying less attention to higher-arity relations although they are ubiquitous in real-world KGs. This paper considers link prediction upon n-ary relational facts and proposes a graph-based approach to this task. The key to our approach is to represent the n-ary structure of a fact as a small heterogeneous graph, and model this graph with edge-biased fully-connected attention. The fully-connected attention captures universal inter-vertex interactions, while with edge-aware attentive biases to particularly encode the graph structure and its heterogeneity. In this fashion, our approach fully models global and local dependencies in each n-ary fact, and hence can more effectively capture associations therein. Extensive evaluation verifies the effectiveness and superiority of our approach. It performs substantially and consistently better than current state-of-the-art across a variety of n-ary relational benchmarks. Our code is publicly available.
Entity Structure Within and Throughout: Modeling Mention Dependencies for Document-Level Relation Extraction
Feb 20, 2021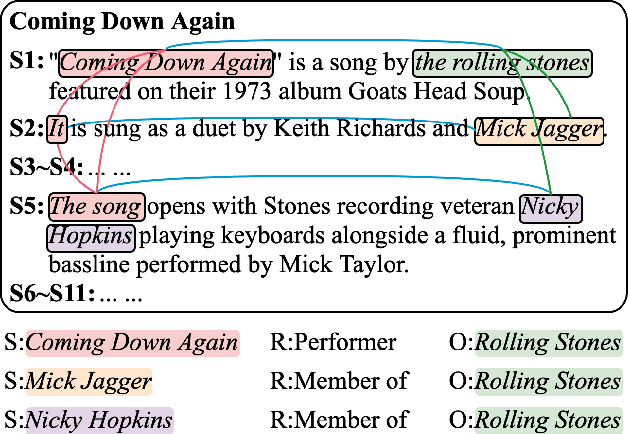


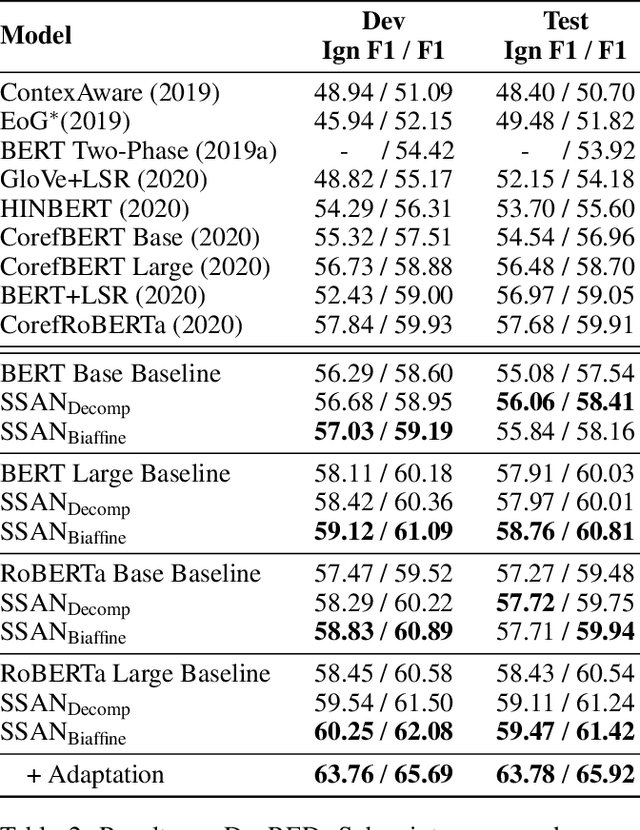
Abstract:Entities, as the essential elements in relation extraction tasks, exhibit certain structure. In this work, we formulate such structure as distinctive dependencies between mention pairs. We then propose SSAN, which incorporates these structural dependencies within the standard self-attention mechanism and throughout the overall encoding stage. Specifically, we design two alternative transformation modules inside each self-attention building block to produce attentive biases so as to adaptively regularize its attention flow. Our experiments demonstrate the usefulness of the proposed entity structure and the effectiveness of SSAN. It significantly outperforms competitive baselines, achieving new state-of-the-art results on three popular document-level relation extraction datasets. We further provide ablation and visualization to show how the entity structure guides the model for better relation extraction. Our code is publicly available.
CoKE: Contextualized Knowledge Graph Embedding
Nov 06, 2019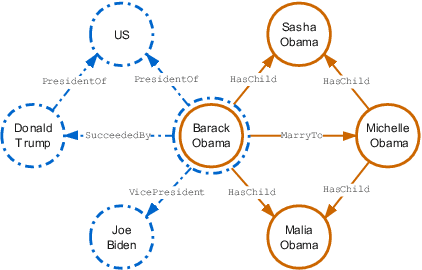
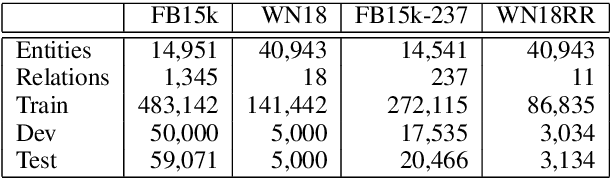
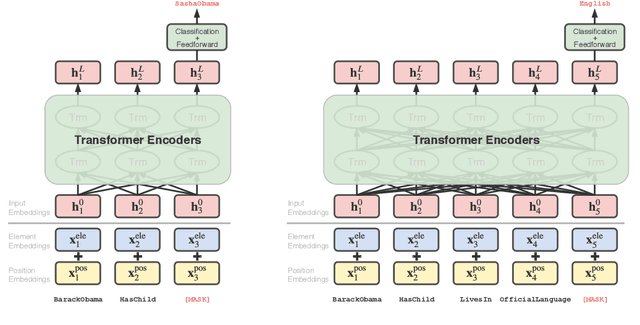
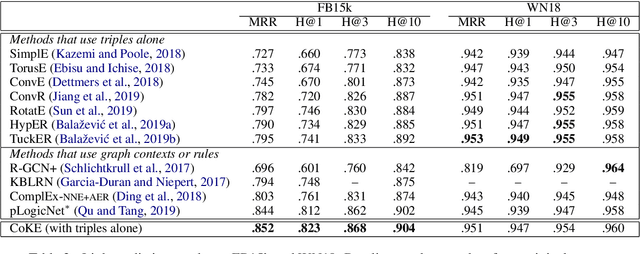
Abstract:Knowledge graph embedding, which projects symbolic entities and relations into continuous vector spaces, is gaining increasing attention. Previous methods allow a single static embedding for each entity or relation, ignoring their intrinsic contextual nature, i.e., entities and relations may appear in different graph contexts, and accordingly, exhibit different properties. This work presents Contextualized Knowledge Graph Embedding (CoKE), a novel paradigm that takes into account such contextual nature, and learns dynamic, flexible, and fully contextualized entity and relation embeddings. Two types of graph contexts are studied: edges and paths, both formulated as sequences of entities and relations. CoKE takes a sequence as input and uses a Transformer encoder to obtain contextualized representations. These representations are hence naturally adaptive to the input, capturing contextual meanings of entities and relations therein. Evaluation on a wide variety of public benchmarks verifies the superiority of CoKE in link prediction and path query answering. It performs consistently better than, or at least equally well as current state-of-the-art in almost every case, in particular offering an absolute improvement of 19.7% in H@10 on path query answering. Our code is available at \url{https://github.com/paddlepaddle/models/tree/develop/PaddleKG/CoKE}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge