Songtai Dai
Building Chinese Biomedical Language Models via Multi-Level Text Discrimination
Oct 14, 2021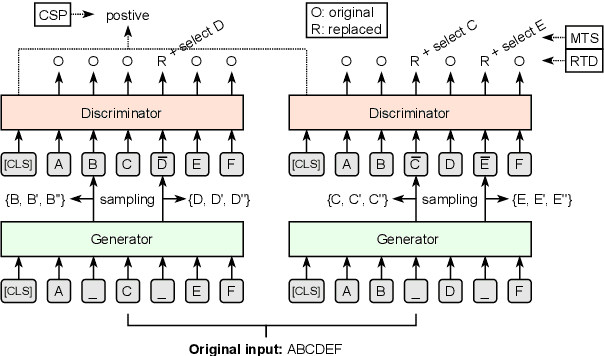
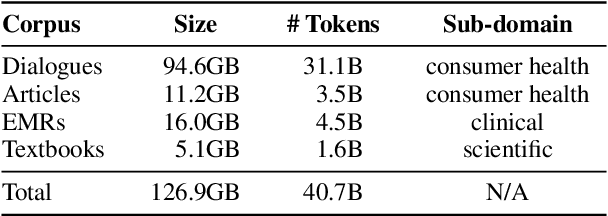
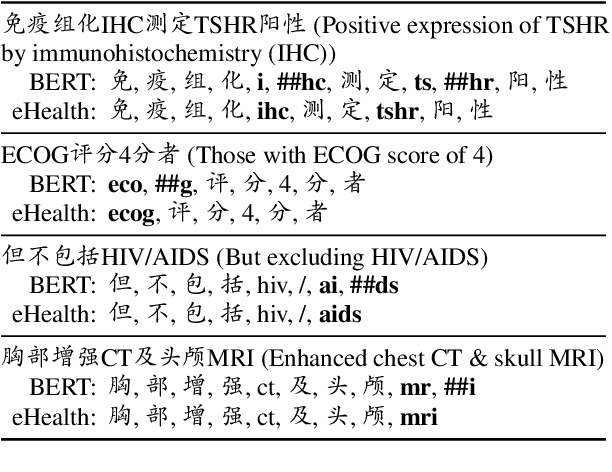
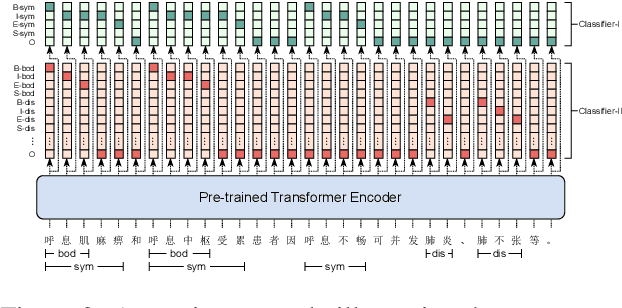
Abstract:Pre-trained language models (PLMs), such as BERT and GPT, have revolutionized the field of NLP, not only in the general domain but also in the biomedical domain. Most prior efforts in building biomedical PLMs have resorted simply to domain adaptation and focused mainly on English. In this work we introduce eHealth, a biomedical PLM in Chinese built with a new pre-training framework. This new framework trains eHealth as a discriminator through both token-level and sequence-level discrimination. The former is to detect input tokens corrupted by a generator and select their original signals from plausible candidates, while the latter is to further distinguish corruptions of a same original sequence from those of the others. As such, eHealth can learn language semantics at both the token and sequence levels. Extensive experiments on 11 Chinese biomedical language understanding tasks of various forms verify the effectiveness and superiority of our approach. The pre-trained model is available to the public at \url{https://github.com/PaddlePaddle/Research/tree/master/KG/eHealth} and the code will also be released later.
CoKE: Contextualized Knowledge Graph Embedding
Nov 06, 2019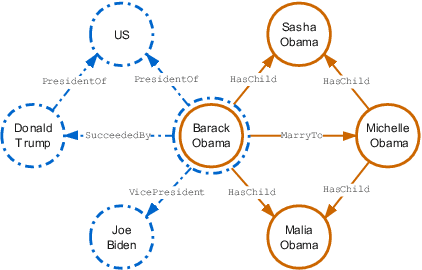
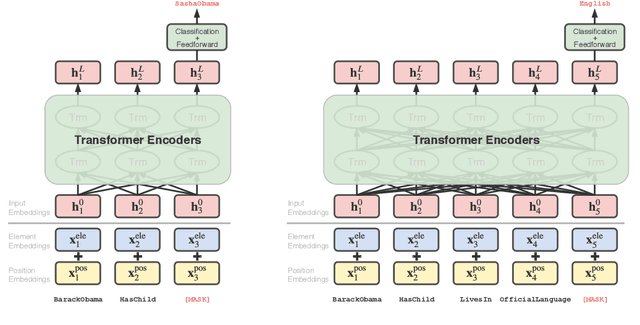
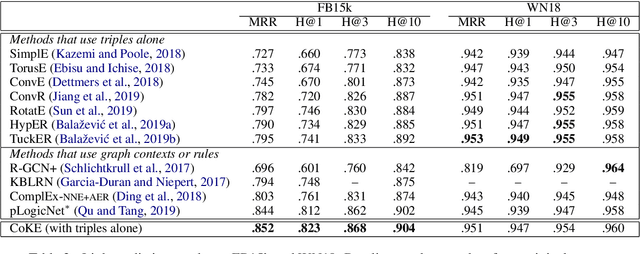
Abstract:Knowledge graph embedding, which projects symbolic entities and relations into continuous vector spaces, is gaining increasing attention. Previous methods allow a single static embedding for each entity or relation, ignoring their intrinsic contextual nature, i.e., entities and relations may appear in different graph contexts, and accordingly, exhibit different properties. This work presents Contextualized Knowledge Graph Embedding (CoKE), a novel paradigm that takes into account such contextual nature, and learns dynamic, flexible, and fully contextualized entity and relation embeddings. Two types of graph contexts are studied: edges and paths, both formulated as sequences of entities and relations. CoKE takes a sequence as input and uses a Transformer encoder to obtain contextualized representations. These representations are hence naturally adaptive to the input, capturing contextual meanings of entities and relations therein. Evaluation on a wide variety of public benchmarks verifies the superiority of CoKE in link prediction and path query answering. It performs consistently better than, or at least equally well as current state-of-the-art in almost every case, in particular offering an absolute improvement of 19.7% in H@10 on path query answering. Our code is available at \url{https://github.com/paddlepaddle/models/tree/develop/PaddleKG/CoKE}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge