Sam Ade Jacobs
Training Ultra Long Context Language Model with Fully Pipelined Distributed Transformer
Aug 30, 2024
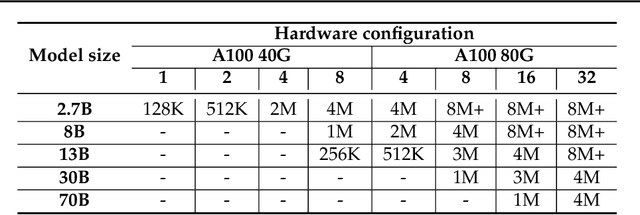
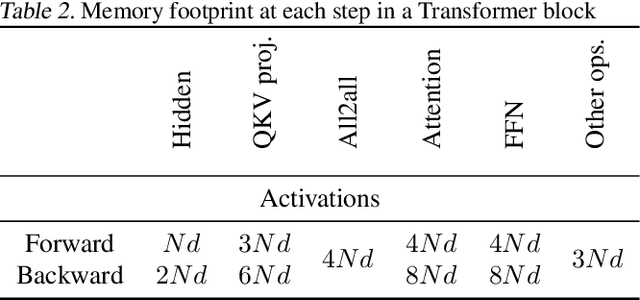

Abstract:Large Language Models (LLMs) with long context capabilities are integral to complex tasks in natural language processing and computational biology, such as text generation and protein sequence analysis. However, training LLMs directly on extremely long contexts demands considerable GPU resources and increased memory, leading to higher costs and greater complexity. Alternative approaches that introduce long context capabilities via downstream finetuning or adaptations impose significant design limitations. In this paper, we propose Fully Pipelined Distributed Transformer (FPDT) for efficiently training long-context LLMs with extreme hardware efficiency. For GPT and Llama models, we achieve a 16x increase in sequence length that can be trained on the same hardware compared to current state-of-the-art solutions. With our dedicated sequence chunk pipeline design, we can now train 8B LLM with 2 million sequence length on only 4 GPUs, while also maintaining over 55% of MFU. Our proposed FPDT is agnostic to existing training techniques and is proven to work efficiently across different LLM models.
Universal Checkpointing: Efficient and Flexible Checkpointing for Large Scale Distributed Training
Jun 27, 2024



Abstract:Existing checkpointing approaches seem ill-suited for distributed training even though hardware limitations make model parallelism, i.e., sharding model state across multiple accelerators, a requirement for model scaling. Consolidating distributed model state into a single checkpoint unacceptably slows down training, and is impractical at extreme scales. Distributed checkpoints, in contrast, are tightly coupled to the model parallelism and hardware configurations of the training run, and thus unusable on different configurations. To address this problem, we propose Universal Checkpointing, a technique that enables efficient checkpoint creation while providing the flexibility of resuming on arbitrary parallelism strategy and hardware configurations. Universal Checkpointing unlocks unprecedented capabilities for large-scale training such as improved resilience to hardware failures through continued training on remaining healthy hardware, and reduced training time through opportunistic exploitation of elastic capacity. The key insight of Universal Checkpointing is the selection of the optimal representation in each phase of the checkpointing life cycle: distributed representation for saving, and consolidated representation for loading. This is achieved using two key mechanisms. First, the universal checkpoint format, which consists of a consolidated representation of each model parameter and metadata for mapping parameter fragments into training ranks of arbitrary model-parallelism configuration. Second, the universal checkpoint language, a simple but powerful specification language for converting distributed checkpoints into the universal checkpoint format. Our evaluation demonstrates the effectiveness and generality of Universal Checkpointing on state-of-the-art model architectures and a wide range of parallelism techniques.
Phi-3 Technical Report: A Highly Capable Language Model Locally on Your Phone
Apr 23, 2024
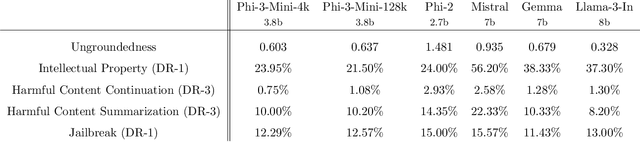
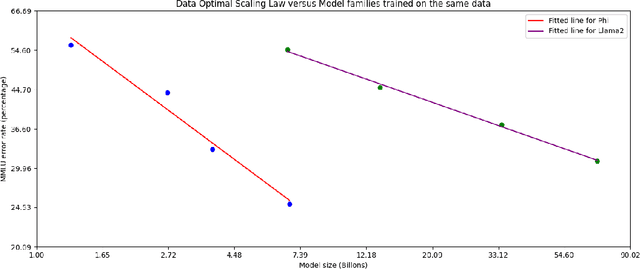
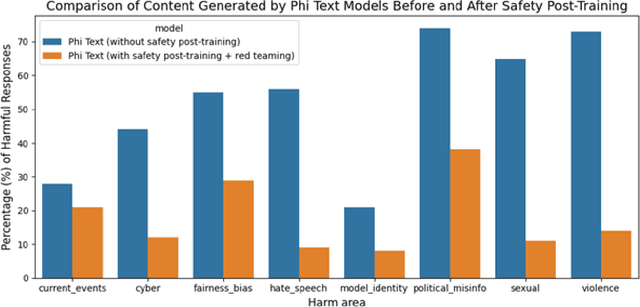
Abstract:We introduce phi-3-mini, a 3.8 billion parameter language model trained on 3.3 trillion tokens, whose overall performance, as measured by both academic benchmarks and internal testing, rivals that of models such as Mixtral 8x7B and GPT-3.5 (e.g., phi-3-mini achieves 69% on MMLU and 8.38 on MT-bench), despite being small enough to be deployed on a phone. The innovation lies entirely in our dataset for training, a scaled-up version of the one used for phi-2, composed of heavily filtered web data and synthetic data. The model is also further aligned for robustness, safety, and chat format. We also provide some initial parameter-scaling results with a 7B and 14B models trained for 4.8T tokens, called phi-3-small and phi-3-medium, both significantly more capable than phi-3-mini (e.g., respectively 75% and 78% on MMLU, and 8.7 and 8.9 on MT-bench).
DeepSpeed Ulysses: System Optimizations for Enabling Training of Extreme Long Sequence Transformer Models
Sep 25, 2023



Abstract:Computation in a typical Transformer-based large language model (LLM) can be characterized by batch size, hidden dimension, number of layers, and sequence length. Until now, system works for accelerating LLM training have focused on the first three dimensions: data parallelism for batch size, tensor parallelism for hidden size and pipeline parallelism for model depth or layers. These widely studied forms of parallelism are not targeted or optimized for long sequence Transformer models. Given practical application needs for long sequence LLM, renewed attentions are being drawn to sequence parallelism. However, existing works in sequence parallelism are constrained by memory-communication inefficiency, limiting their scalability to long sequence large models. In this work, we introduce DeepSpeed-Ulysses, a novel, portable and effective methodology for enabling highly efficient and scalable LLM training with extremely long sequence length. DeepSpeed-Ulysses at its core partitions input data along the sequence dimension and employs an efficient all-to-all collective communication for attention computation. Theoretical communication analysis shows that whereas other methods incur communication overhead as sequence length increases, DeepSpeed-Ulysses maintains constant communication volume when sequence length and compute devices are increased proportionally. Furthermore, experimental evaluations show that DeepSpeed-Ulysses trains 2.5X faster with 4X longer sequence length than the existing method SOTA baseline.
ZeRO++: Extremely Efficient Collective Communication for Giant Model Training
Jun 16, 2023



Abstract:Zero Redundancy Optimizer (ZeRO) has been used to train a wide range of large language models on massive GPUs clusters due to its ease of use, efficiency, and good scalability. However, when training on low-bandwidth clusters, or at scale which forces batch size per GPU to be small, ZeRO's effective throughput is limited because of high communication volume from gathering weights in forward pass, backward pass, and averaging gradients. This paper introduces three communication volume reduction techniques, which we collectively refer to as ZeRO++, targeting each of the communication collectives in ZeRO. First is block-quantization based all-gather. Second is data remapping that trades-off communication for more memory. Third is a novel all-to-all based quantized gradient averaging paradigm as replacement of reduce-scatter collective, which preserves accuracy despite communicating low precision data. Collectively, ZeRO++ reduces communication volume of ZeRO by 4x, enabling up to 2.16x better throughput at 384 GPU scale.
Learning Interpretable Models Through Multi-Objective Neural Architecture Search
Dec 16, 2021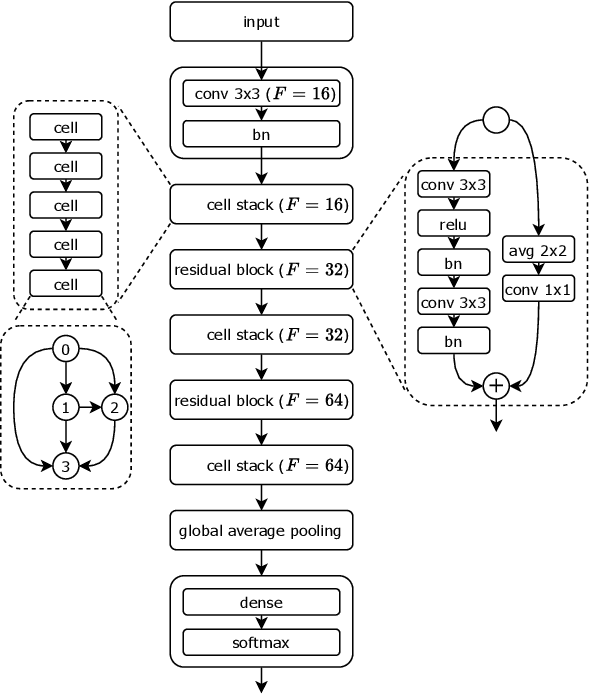
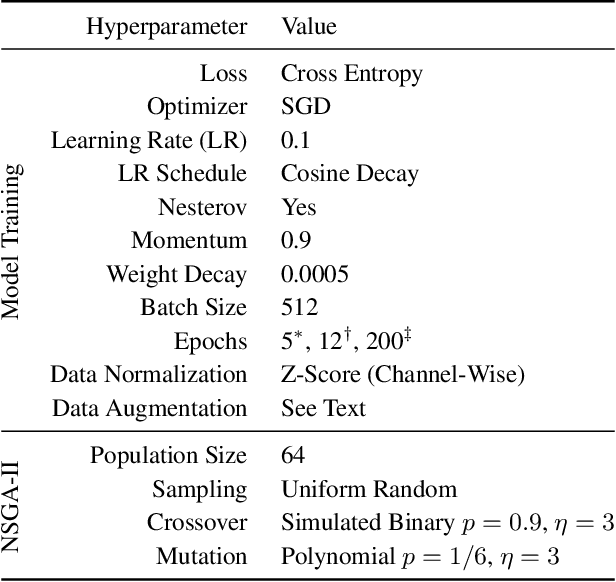
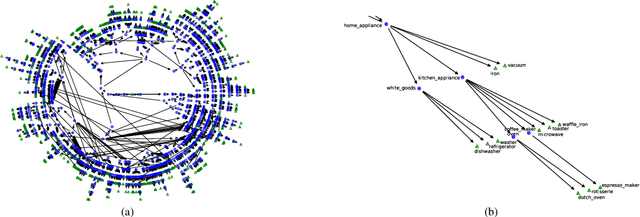
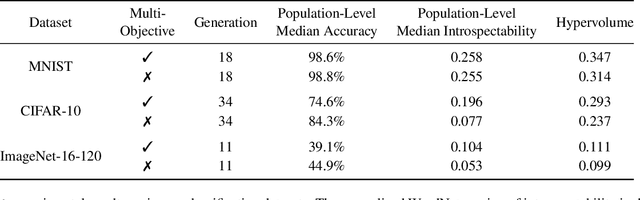
Abstract:Monumental advances in deep learning have led to unprecedented achievements across a multitude of domains. While the performance of deep neural networks is indubitable, the architectural design and interpretability of such models are nontrivial. Research has been introduced to automate the design of neural network architectures through neural architecture search (NAS). Recent progress has made these methods more pragmatic by exploiting distributed computation and novel optimization algorithms. However, there is little work in optimizing architectures for interpretability. To this end, we propose a multi-objective distributed NAS framework that optimizes for both task performance and introspection. We leverage the non-dominated sorting genetic algorithm (NSGA-II) and explainable AI (XAI) techniques to reward architectures that can be better comprehended by humans. The framework is evaluated on several image classification datasets. We demonstrate that jointly optimizing for introspection ability and task error leads to more disentangled architectures that perform within tolerable error.
Merlin: Enabling Machine Learning-Ready HPC Ensembles
Dec 05, 2019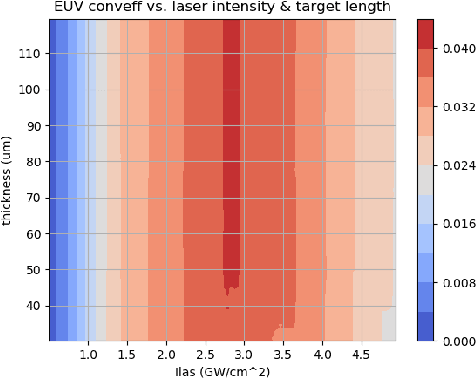
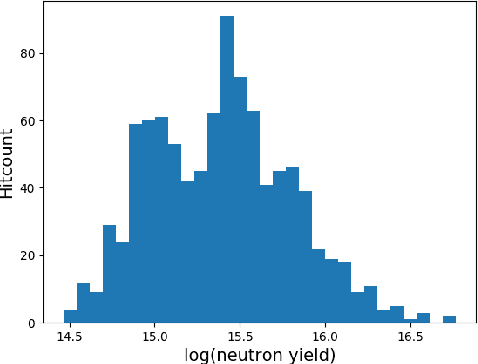
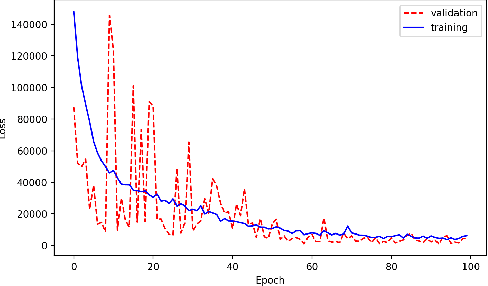
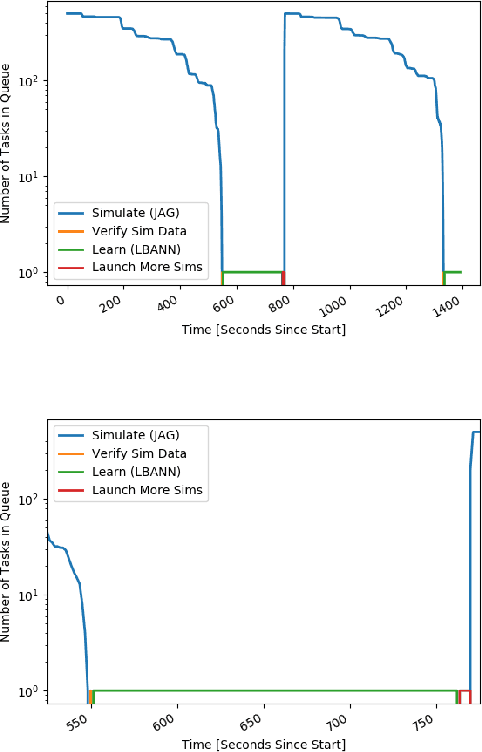
Abstract:With the growing complexity of computational and experimental facilities, many scientific researchers are turning to machine learning (ML) techniques to analyze large scale ensemble data. With complexities such as multi-component workflows, heterogeneous machine architectures, parallel file systems, and batch scheduling, care must be taken to facilitate this analysis in a high performance computing (HPC) environment. In this paper, we present Merlin, a workflow framework to enable large ML-friendly ensembles of scientific HPC simulations. By augmenting traditional HPC with distributed compute technologies, Merlin aims to lower the barrier for scientific subject matter experts to incorporate ML into their analysis. In addition to its design and some examples, we describe how Merlin was deployed on the Sierra Supercomputer at Lawrence Livermore National Laboratory to create an unprecedented benchmark inertial confinement fusion dataset of approximately 100 million individual simulations and over 24 terabytes of multi-modal physics-based scalar, vector and hyperspectral image data.
Parallelizing Training of Deep Generative Models on Massive Scientific Datasets
Oct 05, 2019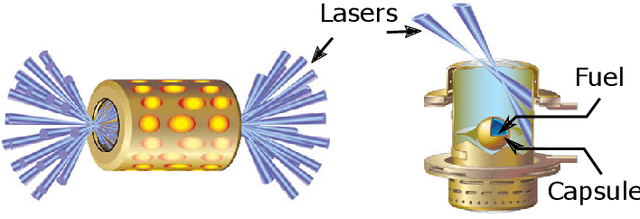
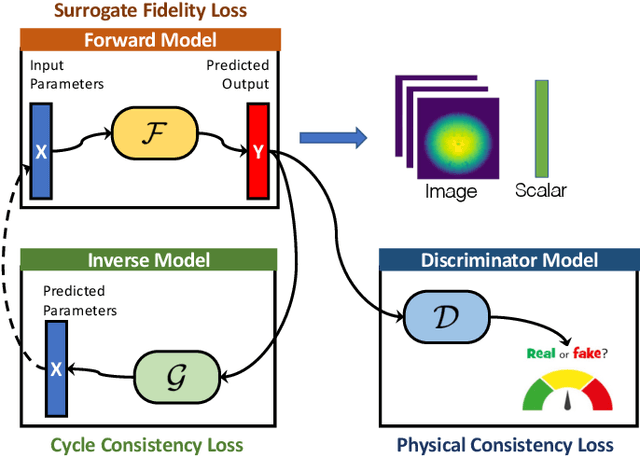
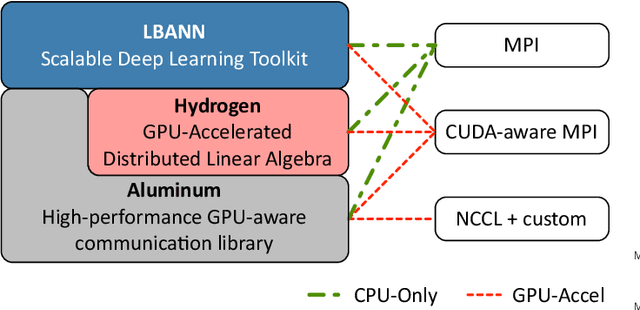
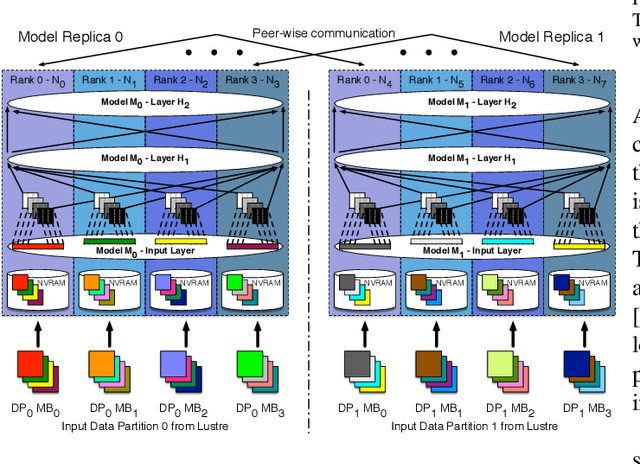
Abstract:Training deep neural networks on large scientific data is a challenging task that requires enormous compute power, especially if no pre-trained models exist to initialize the process. We present a novel tournament method to train traditional as well as generative adversarial networks built on LBANN, a scalable deep learning framework optimized for HPC systems. LBANN combines multiple levels of parallelism and exploits some of the worlds largest supercomputers. We demonstrate our framework by creating a complex predictive model based on multi-variate data from high-energy-density physics containing hundreds of millions of images and hundreds of millions of scalar values derived from tens of millions of simulations of inertial confinement fusion. Our approach combines an HPC workflow and extends LBANN with optimized data ingestion and the new tournament-style training algorithm to produce a scalable neural network architecture using a CORAL-class supercomputer. Experimental results show that 64 trainers (1024 GPUs) achieve a speedup of 70.2 over a single trainer (16 GPUs) baseline, and an effective 109% parallel efficiency.
Scalable Topological Data Analysis and Visualization for Evaluating Data-Driven Models in Scientific Applications
Jul 19, 2019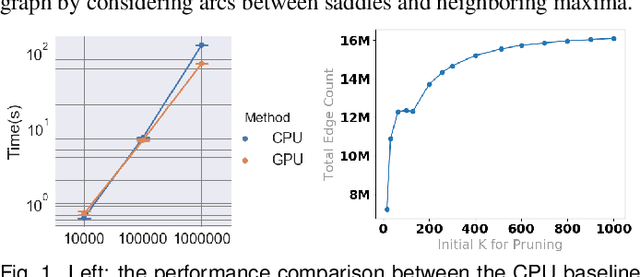
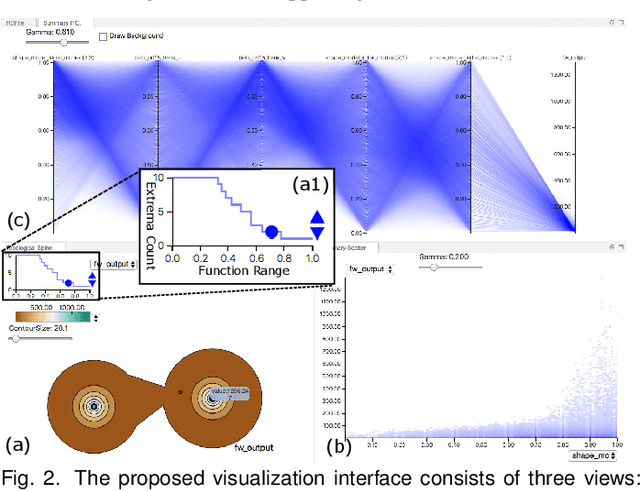
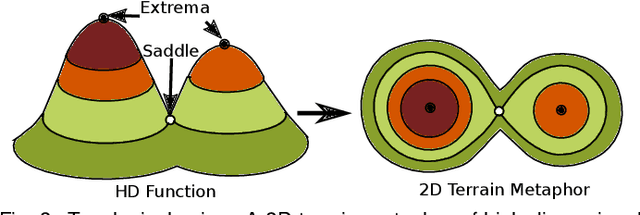
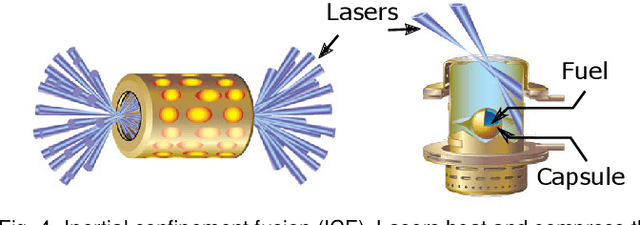
Abstract:With the rapid adoption of machine learning techniques for large-scale applications in science and engineering comes the convergence of two grand challenges in visualization. First, the utilization of black box models (e.g., deep neural networks) calls for advanced techniques in exploring and interpreting model behaviors. Second, the rapid growth in computing has produced enormous datasets that require techniques that can handle millions or more samples. Although some solutions to these interpretability challenges have been proposed, they typically do not scale beyond thousands of samples, nor do they provide the high-level intuition scientists are looking for. Here, we present the first scalable solution to explore and analyze high-dimensional functions often encountered in the scientific data analysis pipeline. By combining a new streaming neighborhood graph construction, the corresponding topology computation, and a novel data aggregation scheme, namely topology aware datacubes, we enable interactive exploration of both the topological and the geometric aspect of high-dimensional data. Following two use cases from high-energy-density (HED) physics and computational biology, we demonstrate how these capabilities have led to crucial new insights in both applications.
Distinguishing between Normal and Cancer Cells Using Autoencoder Node Saliency
Jan 30, 2019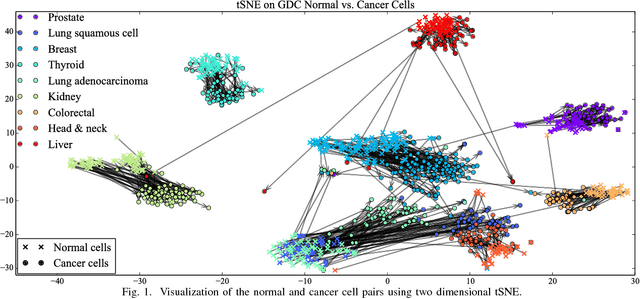
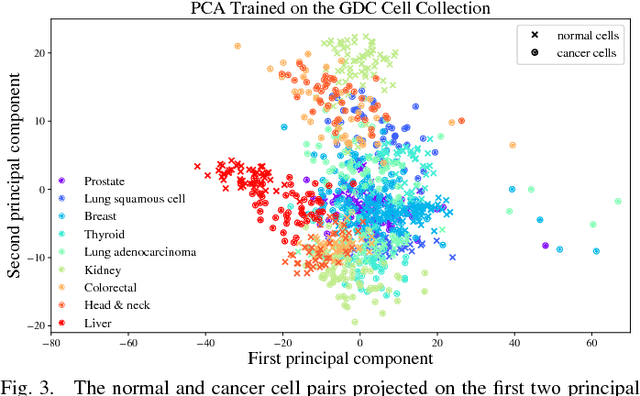
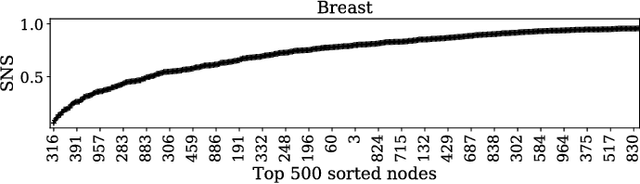
Abstract:Gene expression profiles have been widely used to characterize patterns of cellular responses to diseases. As data becomes available, scalable learning toolkits become essential to processing large datasets using deep learning models to model complex biological processes. We present an autoencoder to capture nonlinear relationships recovered from gene expression profiles. The autoencoder is a nonlinear dimension reduction technique using an artificial neural network, which learns hidden representations of unlabeled data. We train the autoencoder on a large collection of tumor samples from the National Cancer Institute Genomic Data Commons, and obtain a generalized and unsupervised latent representation. We leverage a HPC-focused deep learning toolkit, Livermore Big Artificial Neural Network (LBANN) to efficiently parallelize the training algorithm, reducing computation times from several hours to a few minutes. With the trained autoencoder, we generate latent representations of a small dataset, containing pairs of normal and cancer cells of various tumor types. A novel measure called autoencoder node saliency (ANS) is introduced to identify the hidden nodes that best differentiate various pairs of cells. We compare our findings of the best classifying nodes with principal component analysis and the visualization of t-distributed stochastic neighbor embedding. We demonstrate that the autoencoder effectively extracts distinct gene features for multiple learning tasks in the dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge