Nianzu Yang
Rethinking Video Tokenization: A Conditioned Diffusion-based Approach
Mar 05, 2025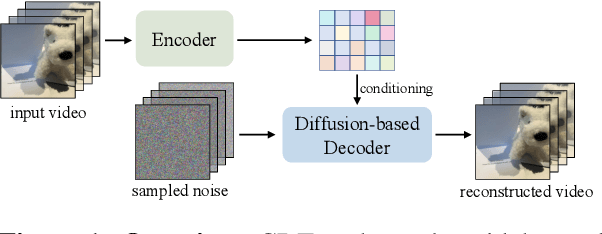
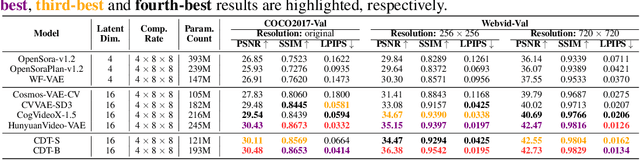
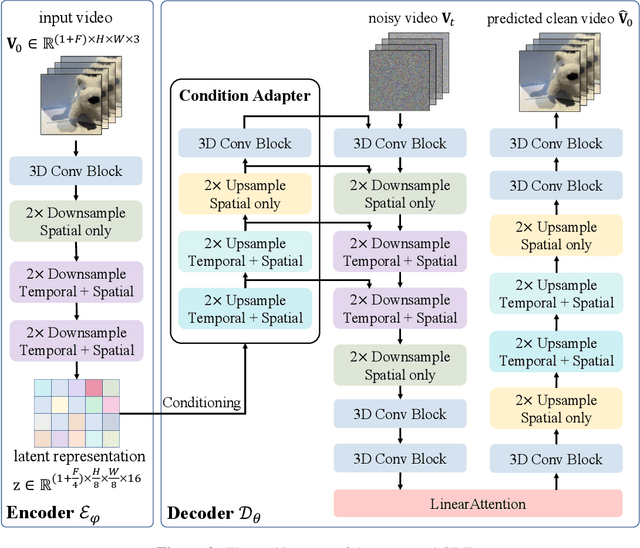
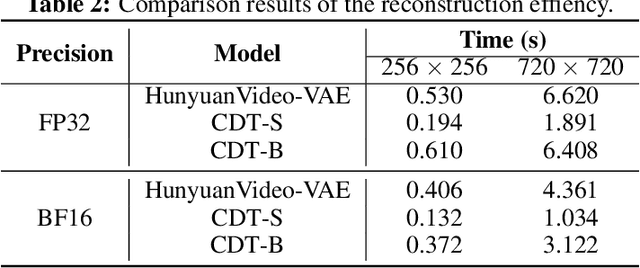
Abstract:Video tokenizers, which transform videos into compact latent representations, are key to video generation. Existing video tokenizers are based on the VAE architecture and follow a paradigm where an encoder compresses videos into compact latents, and a deterministic decoder reconstructs the original videos from these latents. In this paper, we propose a novel \underline{\textbf{C}}onditioned \underline{\textbf{D}}iffusion-based video \underline{\textbf{T}}okenizer entitled \textbf{\ourmethod}, which departs from previous methods by replacing the deterministic decoder with a 3D causal diffusion model. The reverse diffusion generative process of the decoder is conditioned on the latent representations derived via the encoder. With a feature caching and sampling acceleration, the framework efficiently reconstructs high-fidelity videos of arbitrary lengths. Results show that {\ourmethod} achieves state-of-the-art performance in video reconstruction tasks using just a single-step sampling. Even a smaller version of {\ourmethod} still achieves reconstruction results on par with the top two baselines. Furthermore, the latent video generation model trained using {\ourmethod} also shows superior performance.
What Is a Good Caption? A Comprehensive Visual Caption Benchmark for Evaluating Both Correctness and Coverage of MLLMs
Feb 19, 2025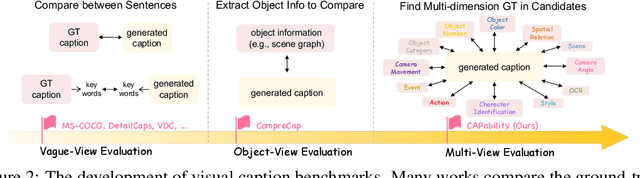
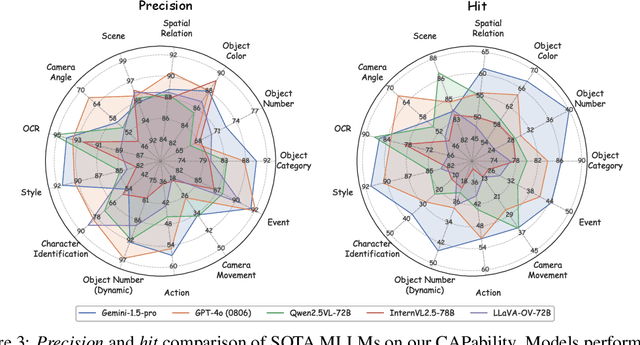

Abstract:Recent advancements in Multimodal Large Language Models (MLLMs) have rendered traditional visual captioning benchmarks obsolete, as they primarily evaluate short descriptions with outdated metrics. While recent benchmarks address these limitations by decomposing captions into visual elements and adopting model-based evaluation, they remain incomplete-overlooking critical aspects, while providing vague, non-explanatory scores. To bridge this gap, we propose CV-CapBench, a Comprehensive Visual Caption Benchmark that systematically evaluates caption quality across 6 views and 13 dimensions. CV-CapBench introduces precision, recall, and hit rate metrics for each dimension, uniquely assessing both correctness and coverage. Experiments on leading MLLMs reveal significant capability gaps, particularly in dynamic and knowledge-intensive dimensions. These findings provide actionable insights for future research. The code and data will be released.
Motion Control for Enhanced Complex Action Video Generation
Nov 13, 2024
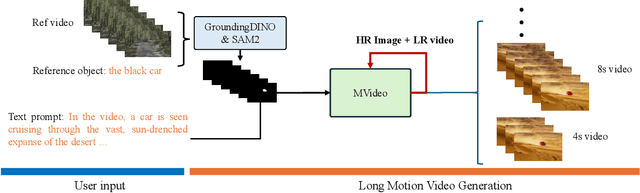

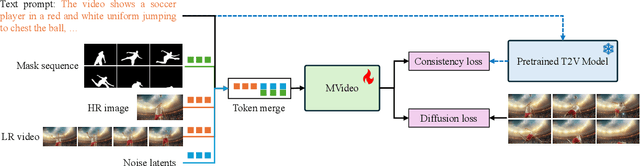
Abstract:Existing text-to-video (T2V) models often struggle with generating videos with sufficiently pronounced or complex actions. A key limitation lies in the text prompt's inability to precisely convey intricate motion details. To address this, we propose a novel framework, MVideo, designed to produce long-duration videos with precise, fluid actions. MVideo overcomes the limitations of text prompts by incorporating mask sequences as an additional motion condition input, providing a clearer, more accurate representation of intended actions. Leveraging foundational vision models such as GroundingDINO and SAM2, MVideo automatically generates mask sequences, enhancing both efficiency and robustness. Our results demonstrate that, after training, MVideo effectively aligns text prompts with motion conditions to produce videos that simultaneously meet both criteria. This dual control mechanism allows for more dynamic video generation by enabling alterations to either the text prompt or motion condition independently, or both in tandem. Furthermore, MVideo supports motion condition editing and composition, facilitating the generation of videos with more complex actions. MVideo thus advances T2V motion generation, setting a strong benchmark for improved action depiction in current video diffusion models. Our project page is available at https://mvideo-v1.github.io/.
A Synchronized Layer-by-layer Growing Approach for Plausible Neuronal Morphology Generation
Jan 17, 2024



Abstract:Neuronal morphology is essential for studying brain functioning and understanding neurodegenerative disorders. As the acquiring of real-world morphology data is expensive, computational approaches especially learning-based ones e.g. MorphVAE for morphology generation were recently studied, which are often conducted in a way of randomly augmenting a given authentic morphology to achieve plausibility. Under such a setting, this paper proposes \textbf{MorphGrower} which aims to generate more plausible morphology samples by mimicking the natural growth mechanism instead of a one-shot treatment as done in MorphVAE. Specifically, MorphGrower generates morphologies layer by layer synchronously and chooses a pair of sibling branches as the basic generation block, and the generation of each layer is conditioned on the morphological structure of previous layers and then generate morphologies via a conditional variational autoencoder with spherical latent space. Extensive experimental results on four real-world datasets demonstrate that MorphGrower outperforms MorphVAE by a notable margin. Our code will be publicly available to facilitate future research.
EasyDGL: Encode, Train and Interpret for Continuous-time Dynamic Graph Learning
Mar 22, 2023Abstract:Dynamic graphs arise in various real-world applications, and it is often welcomed to model the dynamics directly in continuous time domain for its flexibility. This paper aims to design an easy-to-use pipeline (termed as EasyDGL which is also due to its implementation by DGL toolkit) composed of three key modules with both strong fitting ability and interpretability. Specifically the proposed pipeline which involves encoding, training and interpreting: i) a temporal point process (TPP) modulated attention architecture to endow the continuous-time resolution with the coupled spatiotemporal dynamics of the observed graph with edge-addition events; ii) a principled loss composed of task-agnostic TPP posterior maximization based on observed events on the graph, and a task-aware loss with a masking strategy over dynamic graph, where the covered tasks include dynamic link prediction, dynamic node classification and node traffic forecasting; iii) interpretation of the model outputs (e.g., representations and predictions) with scalable perturbation-based quantitative analysis in the graph Fourier domain, which could more comprehensively reflect the behavior of the learned model. Extensive experimental results on public benchmarks show the superior performance of our EasyDGL for time-conditioned predictive tasks, and in particular demonstrate that EasyDGL can effectively quantify the predictive power of frequency content that a model learn from the evolving graph data.
Learning Self-Modulating Attention in Continuous Time Space with Applications to Sequential Recommendation
Mar 30, 2022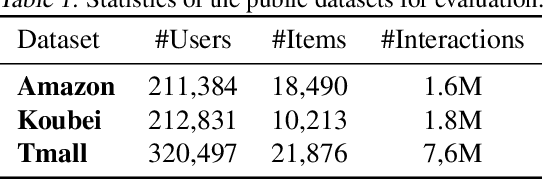
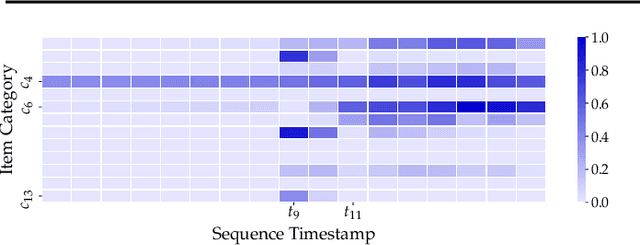
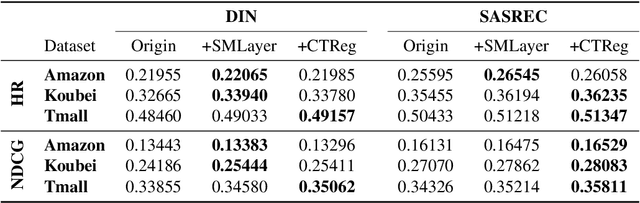
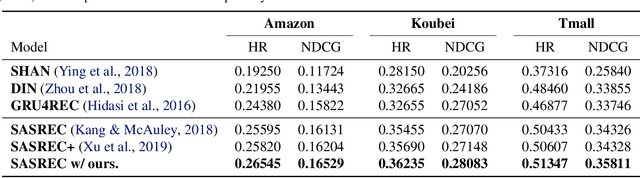
Abstract:User interests are usually dynamic in the real world, which poses both theoretical and practical challenges for learning accurate preferences from rich behavior data. Among existing user behavior modeling solutions, attention networks are widely adopted for its effectiveness and relative simplicity. Despite being extensively studied, existing attentions still suffer from two limitations: i) conventional attentions mainly take into account the spatial correlation between user behaviors, regardless the distance between those behaviors in the continuous time space; and ii) these attentions mostly provide a dense and undistinguished distribution over all past behaviors then attentively encode them into the output latent representations. This is however not suitable in practical scenarios where a user's future actions are relevant to a small subset of her/his historical behaviors. In this paper, we propose a novel attention network, named self-modulating attention, that models the complex and non-linearly evolving dynamic user preferences. We empirically demonstrate the effectiveness of our method on top-N sequential recommendation tasks, and the results on three large-scale real-world datasets show that our model can achieve state-of-the-art performance.
Molecule Generation for Drug Design: a Graph Learning Perspective
Feb 18, 2022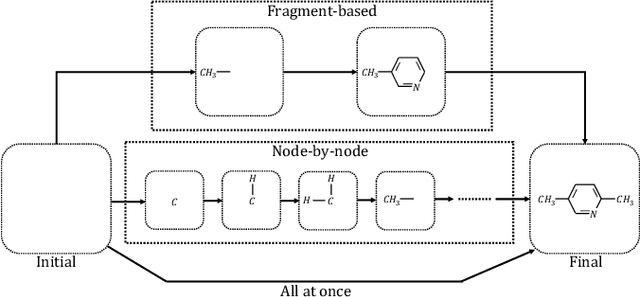
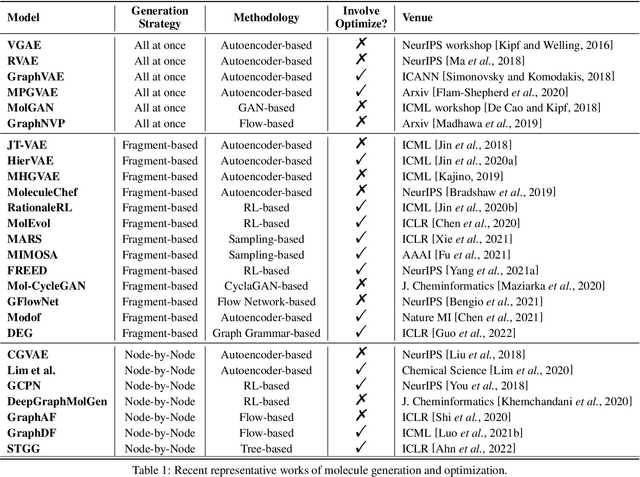
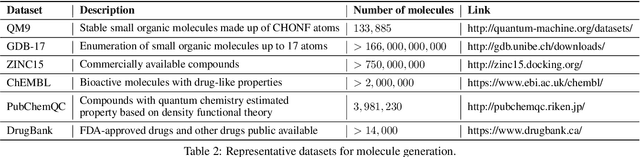
Abstract:Machine learning has revolutionized many fields, and graph learning is recently receiving increasing attention. From the application perspective, one of the emerging and attractive areas is aiding the design and discovery of molecules, especially in drug industry. In this survey, we provide an overview of the state-of-the-art molecule (and mostly for de novo drug) design and discovery aiding methods whose methodology involves (deep) graph learning. Specifically, we propose to categorize these methods into three groups: i) all at once, ii) fragment-based and iii) node-by-node. We further present some representative public datasets and summarize commonly utilized evaluation metrics for generation and optimization, respectively. Finally, we discuss challenges and directions for future research, from the drug design perspective.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge