Mingrui Yang
AROMA: Autonomous Rank-one Matrix Adaptation
Apr 06, 2025



Abstract:As large language models continue to grow in size, parameter-efficient fine-tuning has become increasingly crucial. While low-rank adaptation (LoRA) offers a solution through low-rank updates, its static rank allocation may yield suboptimal results. Adaptive low-rank adaptation (AdaLoRA) improves this with dynamic allocation but remains sensitive to initial and target rank configurations. We introduce AROMA, a framework that automatically constructs layer-specific updates by iteratively building up rank-one components with very few trainable parameters that gradually diminish to zero. Unlike existing methods that employ rank reduction mechanisms, AROMA introduces a dual-loop architecture for rank growth. The inner loop extracts information from each rank-one subspace, while the outer loop determines the number of rank-one subspaces, i.e., the optimal rank. We reset optimizer states to maintain subspace independence. AROMA significantly reduces parameters compared to LoRA and AdaLoRA while achieving superior performance on natural language understanding and commonsense reasoning tasks, offering new insights into adaptive parameter-efficient fine-tuning. The code is available at \href{https://github.com/ShuDun23/AROMA}{AROMA}.
Towards Quantum Tensor Decomposition in Biomedical Applications
Feb 19, 2025


Abstract:Tensor decomposition has emerged as a powerful framework for feature extraction in multi-modal biomedical data. In this review, we present a comprehensive analysis of tensor decomposition methods such as Tucker, CANDECOMP/PARAFAC, spiked tensor decomposition, etc. and their diverse applications across biomedical domains such as imaging, multi-omics, and spatial transcriptomics. To systematically investigate the literature, we applied a topic modeling-based approach that identifies and groups distinct thematic sub-areas in biomedicine where tensor decomposition has been used, thereby revealing key trends and research directions. We evaluated challenges related to the scalability of latent spaces along with obtaining the optimal rank of the tensor, which often hinder the extraction of meaningful features from increasingly large and complex datasets. Additionally, we discuss recent advances in quantum algorithms for tensor decomposition, exploring how quantum computing can be leveraged to address these challenges. Our study includes a preliminary resource estimation analysis for quantum computing platforms and examines the feasibility of implementing quantum-enhanced tensor decomposition methods on near-term quantum devices. Collectively, this review not only synthesizes current applications and challenges of tensor decomposition in biomedical analyses but also outlines promising quantum computing strategies to enhance its impact on deriving actionable insights from complex biomedical data.
Detection-Guided Deep Learning-Based Model with Spatial Regularization for Lung Nodule Segmentation
Oct 26, 2024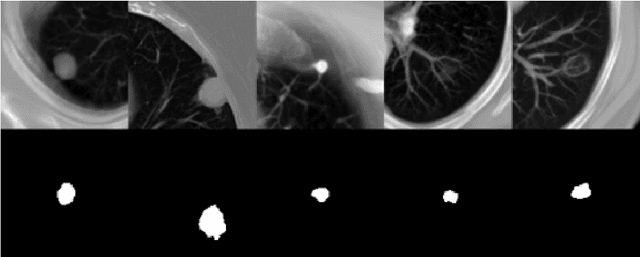
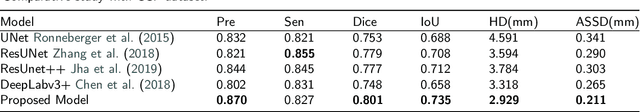
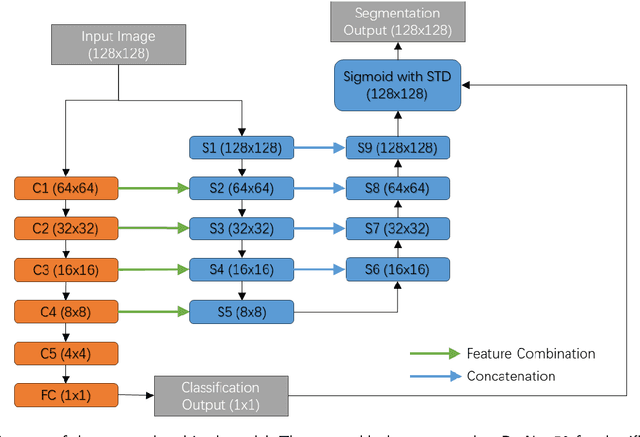
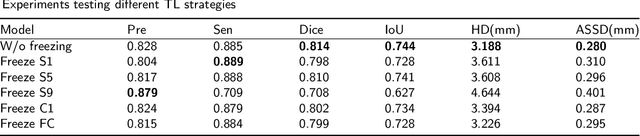
Abstract:Lung cancer ranks as one of the leading causes of cancer diagnosis and is the foremost cause of cancer-related mortality worldwide. The early detection of lung nodules plays a pivotal role in improving outcomes for patients, as it enables timely and effective treatment interventions. The segmentation of lung nodules plays a critical role in aiding physicians in distinguishing between malignant and benign lesions. However, this task remains challenging due to the substantial variation in the shapes and sizes of lung nodules, and their frequent proximity to lung tissues, which complicates clear delineation. In this study, we introduce a novel model for segmenting lung nodules in computed tomography (CT) images, leveraging a deep learning framework that integrates segmentation and classification processes. This model is distinguished by its use of feature combination blocks, which facilitate the sharing of information between the segmentation and classification components. Additionally, we employ the classification outcomes as priors to refine the size estimation of the predicted nodules, integrating these with a spatial regularization technique to enhance precision. Furthermore, recognizing the challenges posed by limited training datasets, we have developed an optimal transfer learning strategy that freezes certain layers to further improve performance. The results show that our proposed model can capture the target nodules more accurately compared to other commonly used models. By applying transfer learning, the performance can be further improved, achieving a sensitivity score of 0.885 and a Dice score of 0.814.
Novel adaptation of video segmentation to 3D MRI: efficient zero-shot knee segmentation with SAM2
Aug 08, 2024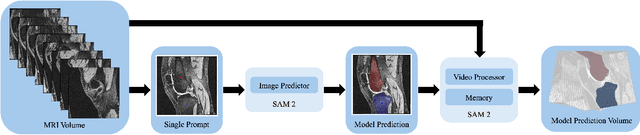
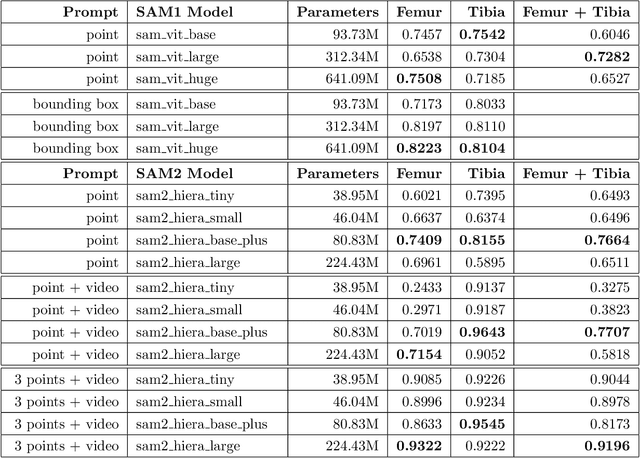
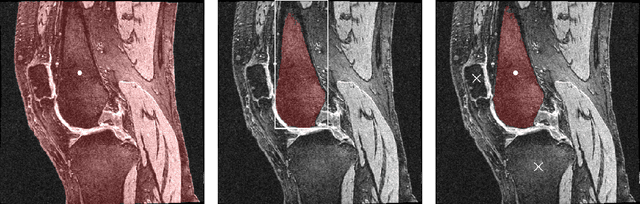
Abstract:Intelligent medical image segmentation methods are rapidly evolving and being increasingly applied, yet they face the challenge of domain transfer, where algorithm performance degrades due to different data distributions between source and target domains. To address this, we introduce a method for zero-shot, single-prompt segmentation of 3D knee MRI by adapting Segment Anything Model 2 (SAM2), a general-purpose segmentation model designed to accept prompts and retain memory across frames of a video. By treating slices from 3D medical volumes as individual video frames, we leverage SAM2's advanced capabilities to generate motion- and spatially-aware predictions. We demonstrate that SAM2 can efficiently perform segmentation tasks in a zero-shot manner with no additional training or fine-tuning, accurately delineating structures in knee MRI scans using only a single prompt. Our experiments on the Osteoarthritis Initiative Zuse Institute Berlin (OAI-ZIB) dataset reveal that SAM2 achieves high accuracy on 3D knee bone segmentation, with a testing Dice similarity coefficient of 0.9643 on tibia. We also present results generated using different SAM2 model sizes, different prompt schemes, as well as comparative results from the SAM1 model deployed on the same dataset. This breakthrough has the potential to revolutionize medical image analysis by providing a scalable, cost-effective solution for automated segmentation, paving the way for broader clinical applications and streamlined workflows.
Interpretable Small Training Set Image Segmentation Network Originated from Multi-Grid Variational Model
Jun 25, 2023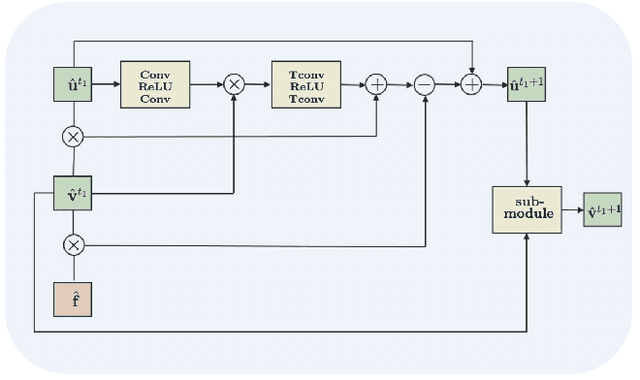
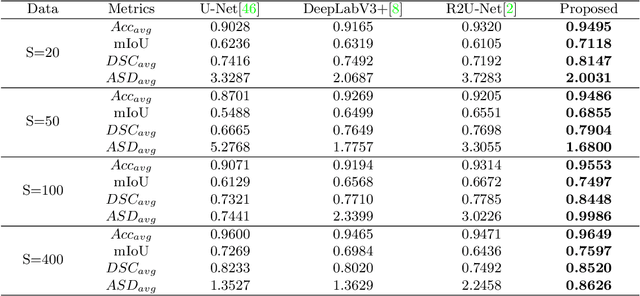
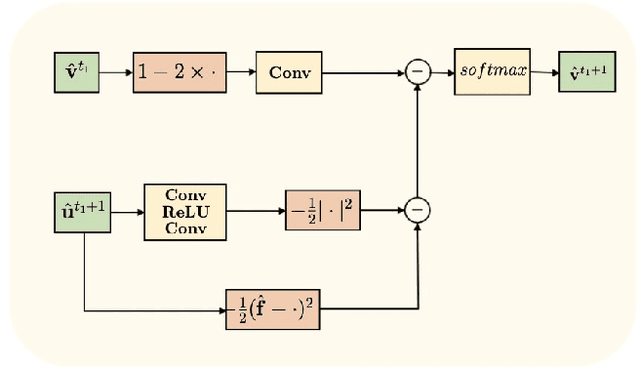
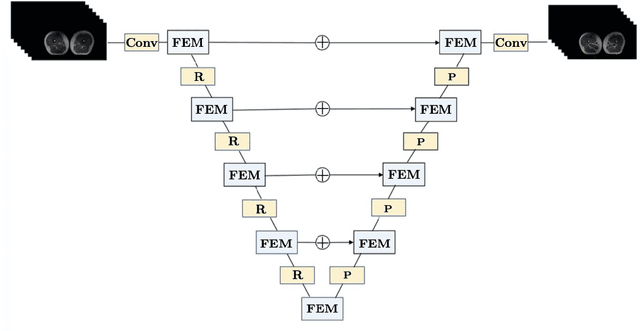
Abstract:The main objective of image segmentation is to divide an image into homogeneous regions for further analysis. This is a significant and crucial task in many applications such as medical imaging. Deep learning (DL) methods have been proposed and widely used for image segmentation. However, these methods usually require a large amount of manually segmented data as training data and suffer from poor interpretability (known as the black box problem). The classical Mumford-Shah (MS) model is effective for segmentation and provides a piece-wise smooth approximation of the original image. In this paper, we replace the hand-crafted regularity term in the MS model with a data adaptive generalized learnable regularity term and use a multi-grid framework to unroll the MS model and obtain a variational model-based segmentation network with better generalizability and interpretability. This approach allows for the incorporation of learnable prior information into the network structure design. Moreover, the multi-grid framework enables multi-scale feature extraction and offers a mathematical explanation for the effectiveness of the U-shaped network structure in producing good image segmentation results. Due to the proposed network originates from a variational model, it can also handle small training sizes. Our experiments on the REFUGE dataset, the White Blood Cell image dataset, and 3D thigh muscle magnetic resonance (MR) images demonstrate that even with smaller training datasets, our method yields better segmentation results compared to related state of the art segmentation methods.
The International Workshop on Osteoarthritis Imaging Knee MRI Segmentation Challenge: A Multi-Institute Evaluation and Analysis Framework on a Standardized Dataset
May 26, 2020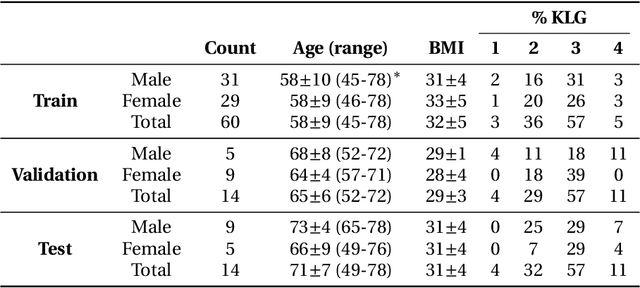
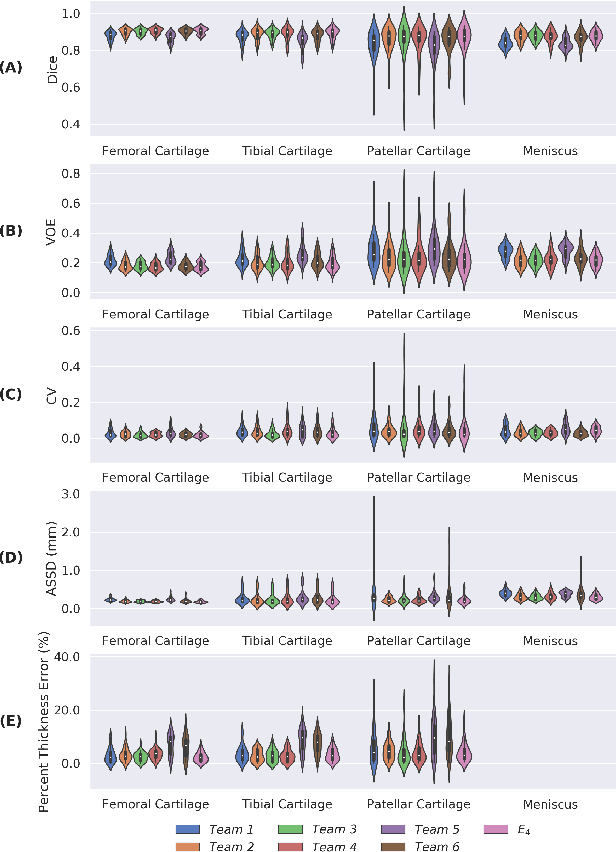
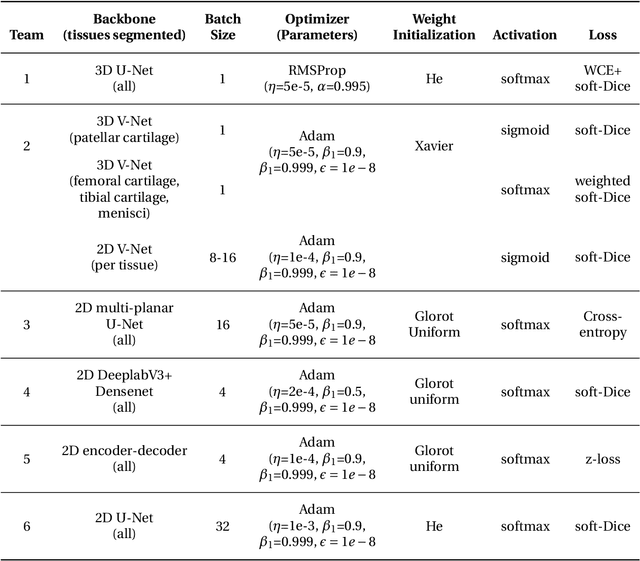
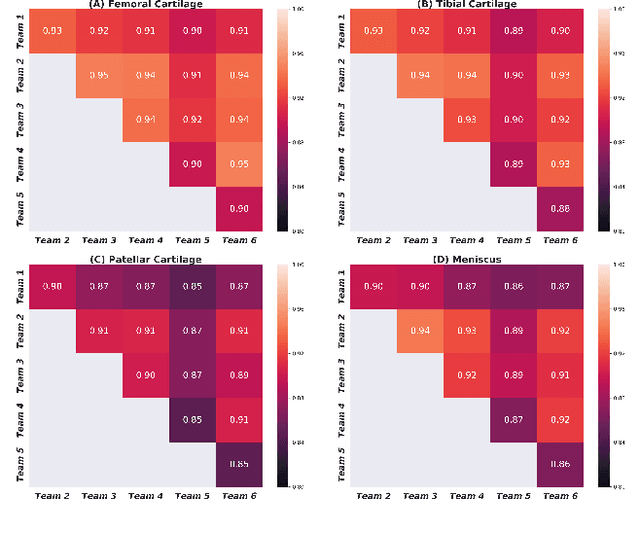
Abstract:Purpose: To organize a knee MRI segmentation challenge for characterizing the semantic and clinical efficacy of automatic segmentation methods relevant for monitoring osteoarthritis progression. Methods: A dataset partition consisting of 3D knee MRI from 88 subjects at two timepoints with ground-truth articular (femoral, tibial, patellar) cartilage and meniscus segmentations was standardized. Challenge submissions and a majority-vote ensemble were evaluated using Dice score, average symmetric surface distance, volumetric overlap error, and coefficient of variation on a hold-out test set. Similarities in network segmentations were evaluated using pairwise Dice correlations. Articular cartilage thickness was computed per-scan and longitudinally. Correlation between thickness error and segmentation metrics was measured using Pearson's coefficient. Two empirical upper bounds for ensemble performance were computed using combinations of model outputs that consolidated true positives and true negatives. Results: Six teams (T1-T6) submitted entries for the challenge. No significant differences were observed across all segmentation metrics for all tissues (p=1.0) among the four top-performing networks (T2, T3, T4, T6). Dice correlations between network pairs were high (>0.85). Per-scan thickness errors were negligible among T1-T4 (p=0.99) and longitudinal changes showed minimal bias (<0.03mm). Low correlations (<0.41) were observed between segmentation metrics and thickness error. The majority-vote ensemble was comparable to top performing networks (p=1.0). Empirical upper bound performances were similar for both combinations (p=1.0). Conclusion: Diverse networks learned to segment the knee similarly where high segmentation accuracy did not correlate to cartilage thickness accuracy. Voting ensembles did not outperform individual networks but may help regularize individual models.
Game of Learning Bloch Equation Simulations for MR Fingerprinting
Apr 05, 2020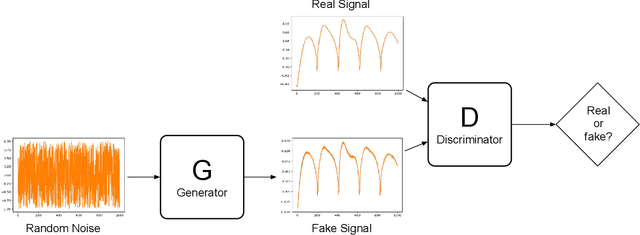
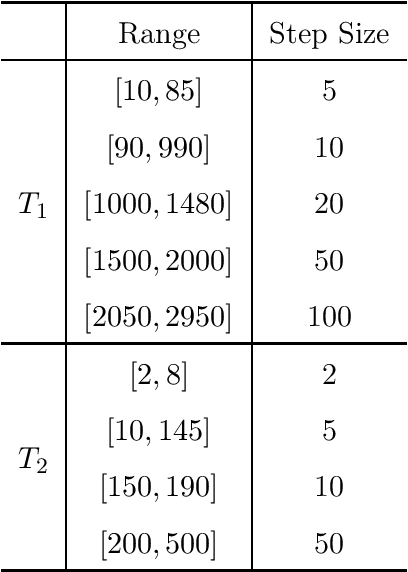
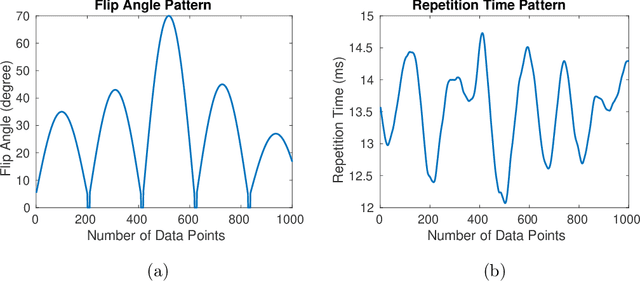
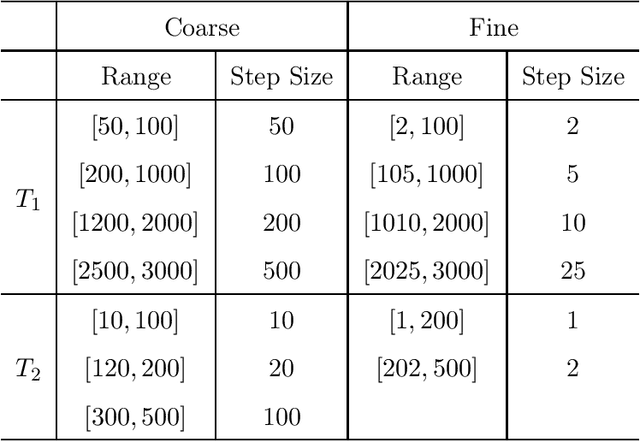
Abstract:Purpose: This work proposes a novel approach to efficiently generate MR fingerprints for MR fingerprinting (MRF) problems based on the unsupervised deep learning model generative adversarial networks (GAN). Methods: The GAN model is adopted and modified for better convergence and performance, resulting in an MRF specific model named GAN-MRF. The GAN-MRF model is trained, validated, and tested using different MRF fingerprints simulated from the Bloch equations with certain MRF sequence. The performance and robustness of the model are further tested by using in vivo data collected on a 3 Tesla scanner from a healthy volunteer together with MRF dictionaries with different sizes. T1, T2 maps are generated and compared quantitatively. Results: The validation and testing curves for the GAN-MRF model show no evidence of high bias or high variance problems. The sample MRF fingerprints generated from the trained GAN-MRF model agree well with the benchmark fingerprints simulated from the Bloch equations. The in vivo T1, T2 maps generated from the GAN-MRF fingerprints are in good agreement with those generated from the Bloch simulated fingerprints, showing good performance and robustness of the proposed GAN-MRF model. Moreover, the MRF dictionary generation time is reduced from hours to sub-second for the testing dictionary. Conclusion: The GAN-MRF model enables a fast and accurate generation of the MRF fingerprints. It significantly reduces the MRF dictionary generation process and opens the door for real-time applications and sequence optimization problems.
Compressive hyperspectral imaging via adaptive sampling and dictionary learning
Dec 02, 2015


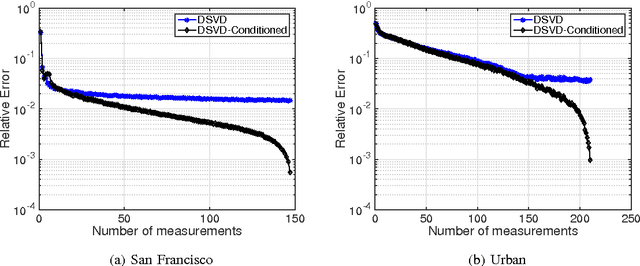
Abstract:In this paper, we propose a new sampling strategy for hyperspectral signals that is based on dictionary learning and singular value decomposition (SVD). Specifically, we first learn a sparsifying dictionary from training spectral data using dictionary learning. We then perform an SVD on the dictionary and use the first few left singular vectors as the rows of the measurement matrix to obtain the compressive measurements for reconstruction. The proposed method provides significant improvement over the conventional compressive sensing approaches. The reconstruction performance is further improved by reconditioning the sensing matrix using matrix balancing. We also demonstrate that the combination of dictionary learning and SVD is robust by applying them to different datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge