Omar Shehab
Towards Quantum Tensor Decomposition in Biomedical Applications
Feb 19, 2025

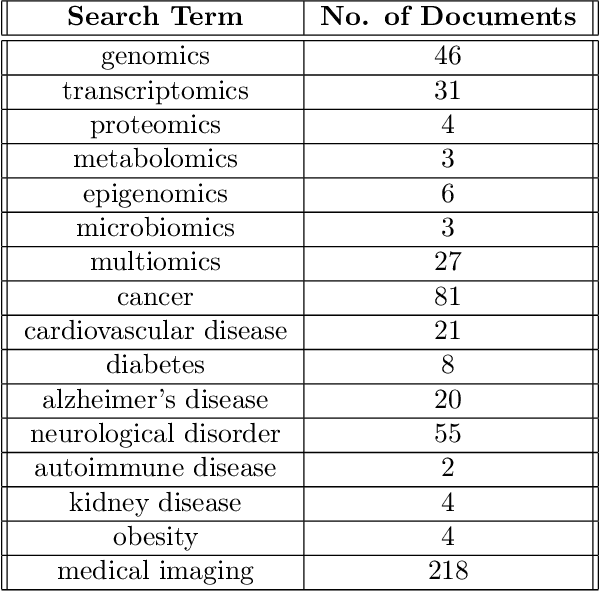

Abstract:Tensor decomposition has emerged as a powerful framework for feature extraction in multi-modal biomedical data. In this review, we present a comprehensive analysis of tensor decomposition methods such as Tucker, CANDECOMP/PARAFAC, spiked tensor decomposition, etc. and their diverse applications across biomedical domains such as imaging, multi-omics, and spatial transcriptomics. To systematically investigate the literature, we applied a topic modeling-based approach that identifies and groups distinct thematic sub-areas in biomedicine where tensor decomposition has been used, thereby revealing key trends and research directions. We evaluated challenges related to the scalability of latent spaces along with obtaining the optimal rank of the tensor, which often hinder the extraction of meaningful features from increasingly large and complex datasets. Additionally, we discuss recent advances in quantum algorithms for tensor decomposition, exploring how quantum computing can be leveraged to address these challenges. Our study includes a preliminary resource estimation analysis for quantum computing platforms and examines the feasibility of implementing quantum-enhanced tensor decomposition methods on near-term quantum devices. Collectively, this review not only synthesizes current applications and challenges of tensor decomposition in biomedical analyses but also outlines promising quantum computing strategies to enhance its impact on deriving actionable insights from complex biomedical data.
Quantum kernels for real-world predictions based on electronic health records
Dec 12, 2021



Abstract:In recent years, research on near-term quantum machine learning has explored how classical machine learning algorithms endowed with access to quantum kernels (similarity measures) can outperform their purely classical counterparts. Although theoretical work has shown provable advantage on synthetic data sets, no work done to date has studied empirically whether quantum advantage is attainable and with what kind of data set. In this paper, we report the first systematic investigation of empirical quantum advantage (EQA) in healthcare and life sciences and propose an end-to-end framework to study EQA. We selected electronic health records (EHRs) data subsets and created a configuration space of 5-20 features and 200-300 training samples. For each configuration coordinate, we trained classical support vector machine (SVM) models based on radial basis function (RBF) kernels and quantum models with custom kernels using an IBM quantum computer. We empirically identified regimes where quantum kernels could provide advantage on a particular data set and introduced a terrain ruggedness index, a metric to help quantitatively estimate how the accuracy of a given model will perform as a function of the number of features and sample size. The generalizable framework introduced here represents a key step towards a priori identification of data sets where quantum advantage could exist.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge