Richard Lartey
Towards Quantum Tensor Decomposition in Biomedical Applications
Feb 19, 2025


Abstract:Tensor decomposition has emerged as a powerful framework for feature extraction in multi-modal biomedical data. In this review, we present a comprehensive analysis of tensor decomposition methods such as Tucker, CANDECOMP/PARAFAC, spiked tensor decomposition, etc. and their diverse applications across biomedical domains such as imaging, multi-omics, and spatial transcriptomics. To systematically investigate the literature, we applied a topic modeling-based approach that identifies and groups distinct thematic sub-areas in biomedicine where tensor decomposition has been used, thereby revealing key trends and research directions. We evaluated challenges related to the scalability of latent spaces along with obtaining the optimal rank of the tensor, which often hinder the extraction of meaningful features from increasingly large and complex datasets. Additionally, we discuss recent advances in quantum algorithms for tensor decomposition, exploring how quantum computing can be leveraged to address these challenges. Our study includes a preliminary resource estimation analysis for quantum computing platforms and examines the feasibility of implementing quantum-enhanced tensor decomposition methods on near-term quantum devices. Collectively, this review not only synthesizes current applications and challenges of tensor decomposition in biomedical analyses but also outlines promising quantum computing strategies to enhance its impact on deriving actionable insights from complex biomedical data.
A Geometric Flow Approach for Segmentation of Images with Inhomongeneous Intensity and Missing Boundaries
Sep 19, 2023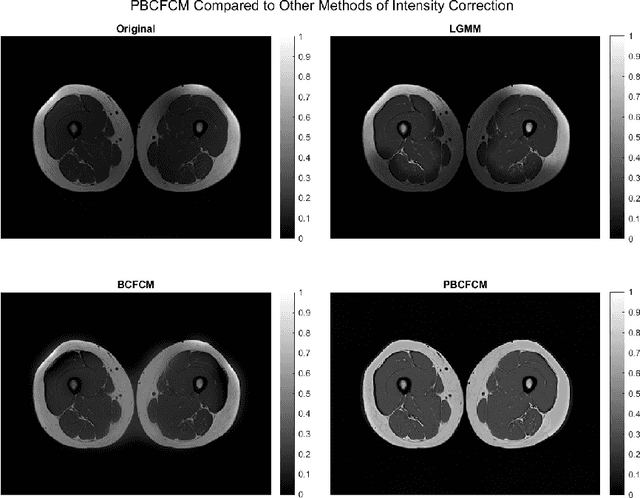
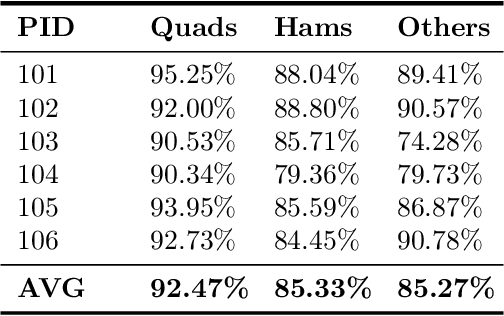
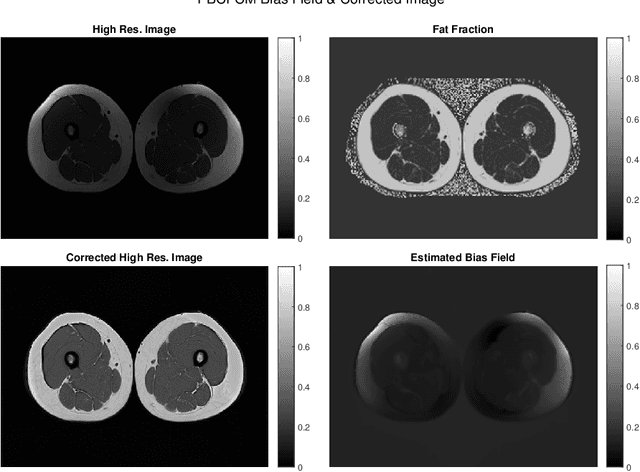

Abstract:Image segmentation is a complex mathematical problem, especially for images that contain intensity inhomogeneity and tightly packed objects with missing boundaries in between. For instance, Magnetic Resonance (MR) muscle images often contain both of these issues, making muscle segmentation especially difficult. In this paper we propose a novel intensity correction and a semi-automatic active contour based segmentation approach. The approach uses a geometric flow that incorporates a reproducing kernel Hilbert space (RKHS) edge detector and a geodesic distance penalty term from a set of markers and anti-markers. We test the proposed scheme on MR muscle segmentation and compare with some state of the art methods. To help deal with the intensity inhomogeneity in this particular kind of image, a new approach to estimate the bias field using a fat fraction image, called Prior Bias-Corrected Fuzzy C-means (PBCFCM), is introduced. Numerical experiments show that the proposed scheme leads to significantly better results than compared ones. The average dice values of the proposed method are 92.5%, 85.3%, 85.3% for quadriceps, hamstrings and other muscle groups while other approaches are at least 10% worse.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge