Maxime Sermesant
EPIONE
Constraint-Based Model in Multimodal Learning to Improve Ventricular Arrhythmia Prediction
Dec 18, 2024Abstract:Cardiac disease evaluation depends on multiple diagnostic modalities: electrocardiogram (ECG) to diagnose abnormal heart rhythms, and imaging modalities such as Magnetic Resonance Imaging (MRI), Computed Tomography (CT) and echocardiography to detect signs of structural abnormalities. Each of these modalities brings complementary information for a better diagnosis of cardiac dysfunction. However, training a machine learning (ML) model with data from multiple modalities is a challenging task, as it increases the dimension space, while keeping constant the number of samples. In fact, as the dimension of the input space increases, the volume of data required for accurate generalisation grows exponentially. In this work, we address this issue, for the application of Ventricular Arrhythmia (VA) prediction, based on the combined clinical and CT imaging features, where we constrained the learning process on medical images (CT) based on the prior knowledge acquired from clinical data. The VA classifier is fed with features extracted from a 3D myocardium thickness map (TM) of the left ventricle. The TM is generated by our pipeline from the imaging input and a Graph Convolutional Network is used as the feature extractor of the 3D TM. We introduce a novel Sequential Fusion method and evaluate its performance against traditional Early Fusion techniques and single-modality models. The crossvalidation results show that the Sequential Fusion model achieved the highest average scores of 80.7% $\pm$ 4.4 Sensitivity and 73.1% $\pm$ 6.0 F1 score, outperforming the Early Fusion model at 65.0% $\pm$ 8.9 Sensitivity and 63.1% $\pm$ 6.3 F1 score. Both fusion models achieved better scores than the single-modality models, where the average Sensitivity and F1 score are 62.8% $\pm$ 10.1; 52.1% $\pm$ 6.5 for the clinical data modality and 62.9% $\pm$ 6.3; 60.7% $\pm$ 5.3 for the medical images modality.
Diffusion based Zero-shot Medical Image-to-Image Translation for Cross Modality Segmentation
Apr 09, 2024

Abstract:Cross-modality image segmentation aims to segment the target modalities using a method designed in the source modality. Deep generative models can translate the target modality images into the source modality, thus enabling cross-modality segmentation. However, a vast body of existing cross-modality image translation methods relies on supervised learning. In this work, we aim to address the challenge of zero-shot learning-based image translation tasks (extreme scenarios in the target modality is unseen in the training phase). To leverage generative learning for zero-shot cross-modality image segmentation, we propose a novel unsupervised image translation method. The framework learns to translate the unseen source image to the target modality for image segmentation by leveraging the inherent statistical consistency between different modalities for diffusion guidance. Our framework captures identical cross-modality features in the statistical domain, offering diffusion guidance without relying on direct mappings between the source and target domains. This advantage allows our method to adapt to changing source domains without the need for retraining, making it highly practical when sufficient labeled source domain data is not available. The proposed framework is validated in zero-shot cross-modality image segmentation tasks through empirical comparisons with influential generative models, including adversarial-based and diffusion-based models.
Zero-shot-Learning Cross-Modality Data Translation Through Mutual Information Guided Stochastic Diffusion
Jan 31, 2023
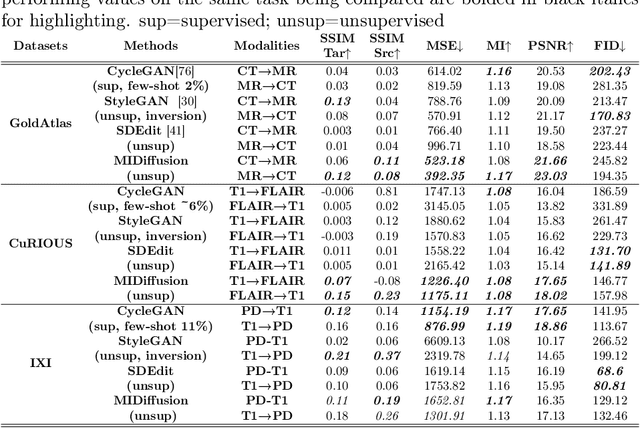
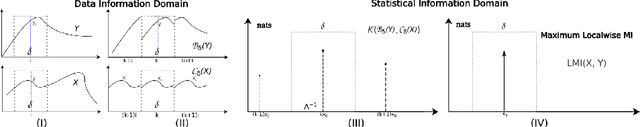
Abstract:Cross-modality data translation has attracted great interest in image computing. Deep generative models (\textit{e.g.}, GANs) show performance improvement in tackling those problems. Nevertheless, as a fundamental challenge in image translation, the problem of Zero-shot-Learning Cross-Modality Data Translation with fidelity remains unanswered. This paper proposes a new unsupervised zero-shot-learning method named Mutual Information guided Diffusion cross-modality data translation Model (MIDiffusion), which learns to translate the unseen source data to the target domain. The MIDiffusion leverages a score-matching-based generative model, which learns the prior knowledge in the target domain. We propose a differentiable local-wise-MI-Layer ($LMI$) for conditioning the iterative denoising sampling. The $LMI$ captures the identical cross-modality features in the statistical domain for the diffusion guidance; thus, our method does not require retraining when the source domain is changed, as it does not rely on any direct mapping between the source and target domains. This advantage is critical for applying cross-modality data translation methods in practice, as a reasonable amount of source domain dataset is not always available for supervised training. We empirically show the advanced performance of MIDiffusion in comparison with an influential group of generative models, including adversarial-based and other score-matching-based models.
Unsupervised Echocardiography Registration through Patch-based MLPs and Transformers
Nov 21, 2022



Abstract:Image registration is an essential but challenging task in medical image computing, especially for echocardiography, where the anatomical structures are relatively noisy compared to other imaging modalities. Traditional (non-learning) registration approaches rely on the iterative optimization of a similarity metric which is usually costly in time complexity. In recent years, convolutional neural network (CNN) based image registration methods have shown good effectiveness. In the meantime, recent studies show that the attention-based model (e.g., Transformer) can bring superior performance in pattern recognition tasks. In contrast, whether the superior performance of the Transformer comes from the long-winded architecture or is attributed to the use of patches for dividing the inputs is unclear yet. This work introduces three patch-based frameworks for image registration using MLPs and transformers. We provide experiments on 2D-echocardiography registration to answer the former question partially and provide a benchmark solution. Our results on a large public 2D echocardiography dataset show that the patch-based MLP/Transformer model can be effectively used for unsupervised echocardiography registration. They demonstrate comparable and even better registration performance than a popular CNN registration model. In particular, patch-based models better preserve volume changes in terms of Jacobian determinants, thus generating robust registration fields with less unrealistic deformation. Our results demonstrate that patch-based learning methods, whether with attention or not, can perform high-performance unsupervised registration tasks with adequate time and space complexity. Our codes are available https://gitlab.inria.fr/epione/mlp\_transformer\_registration
Cardiac Motion Modeling with Parallel Transport and Shape Splines
Feb 17, 2021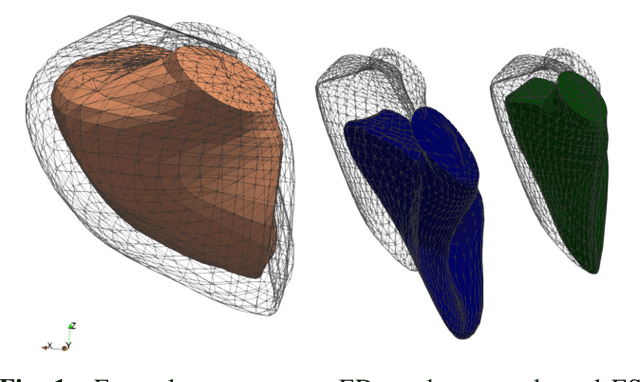
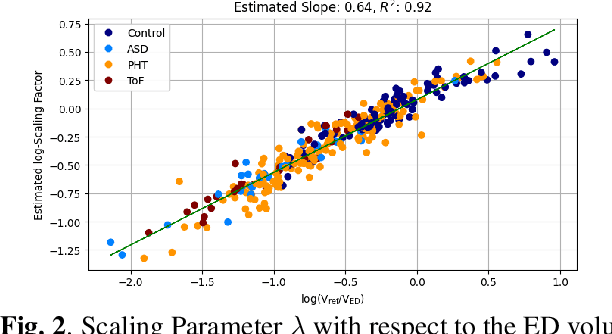
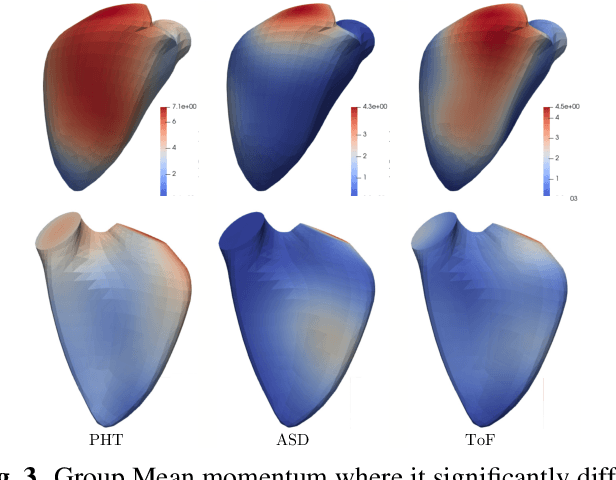
Abstract:In cases of pressure or volume overload, probing cardiac function may be difficult because of the interactions between shape and deformations.In this work, we use the LDDMM framework and parallel transport to estimate and reorient deformations of the right ventricle. We then propose a normalization procedure for the amplitude of the deformation, and a second-order spline model to represent the full cardiac contraction. The method is applied to 3D meshes of the right ventricle extracted from echocardiographic sequences of 314 patients divided into three disease categories and a control group. We find significant differences between pathologies in the model parameters, revealing insights into the dynamics of each disease.
Joint data imputation and mechanistic modelling for simulating heart-brain interactions in incomplete datasets
Oct 08, 2020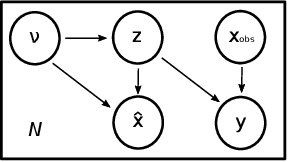
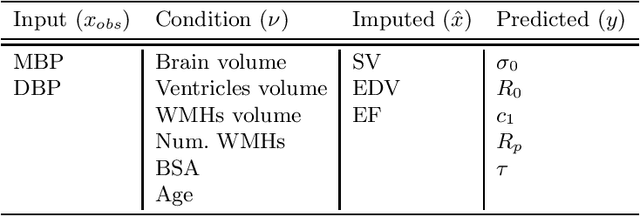
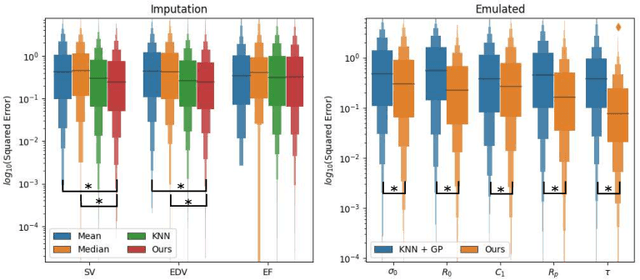
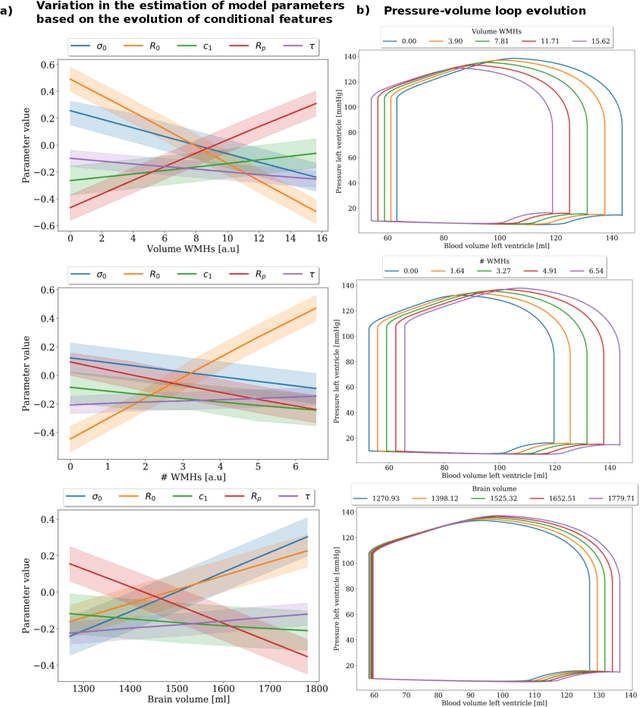
Abstract:The use of mechanistic models in clinical studies is limited by the lack of multi-modal patients data representing different anatomical and physiological processes. For example, neuroimaging datasets do not provide a sufficient representation of heart features for the modeling of cardiovascular factors in brain disorders. To tackle this problem we introduce a probabilistic framework for joint cardiac data imputation and personalisation of cardiovascular mechanistic models, with application to brain studies with incomplete heart data. Our approach is based on a variational framework for the joint inference of an imputation model of cardiac information from the available features, along with a Gaussian Process emulator that can faithfully reproduce personalised cardiovascular dynamics. Experimental results on UK Biobank show that our model allows accurate imputation of missing cardiac features in datasets containing minimal heart information, e.g. systolic and diastolic blood pressures only, while jointly estimating the emulated parameters of the lumped model. This allows a novel exploration of the heart-brain joint relationship through simulation of realistic cardiac dynamics corresponding to different conditions of brain anatomy.
Cardiac Segmentation on Late Gadolinium Enhancement MRI: A Benchmark Study from Multi-Sequence Cardiac MR Segmentation Challenge
Jun 22, 2020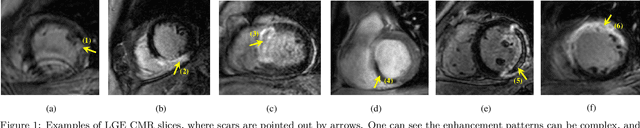
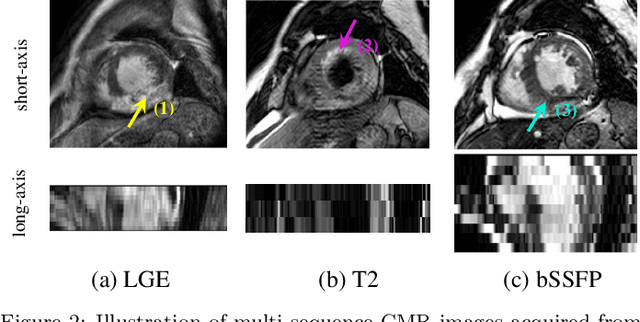
Abstract:Accurate computing, analysis and modeling of the ventricles and myocardium from medical images are important, especially in the diagnosis and treatment management for patients suffering from myocardial infarction (MI). Late gadolinium enhancement (LGE) cardiac magnetic resonance (CMR) provides an important protocol to visualize MI. However, automated segmentation of LGE CMR is still challenging, due to the indistinguishable boundaries, heterogeneous intensity distribution and complex enhancement patterns of pathological myocardium from LGE CMR. Furthermore, compared with the other sequences LGE CMR images with gold standard labels are particularly limited, which represents another obstacle for developing novel algorithms for automatic segmentation of LGE CMR. This paper presents the selective results from the Multi-Sequence Cardiac MR (MS-CMR) Segmentation challenge, in conjunction with MICCAI 2019. The challenge offered a data set of paired MS-CMR images, including auxiliary CMR sequences as well as LGE CMR, from 45 patients who underwent cardiomyopathy. It was aimed to develop new algorithms, as well as benchmark existing ones for LGE CMR segmentation and compare them objectively. In addition, the paired MS-CMR images could enable algorithms to combine the complementary information from the other sequences for the segmentation of LGE CMR. Nine representative works were selected for evaluation and comparisons, among which three methods are unsupervised methods and the other six are supervised. The results showed that the average performance of the nine methods was comparable to the inter-observer variations. The success of these methods was mainly attributed to the inclusion of the auxiliary sequences from the MS-CMR images, which provide important label information for the training of deep neural networks.
A Global Benchmark of Algorithms for Segmenting Late Gadolinium-Enhanced Cardiac Magnetic Resonance Imaging
May 07, 2020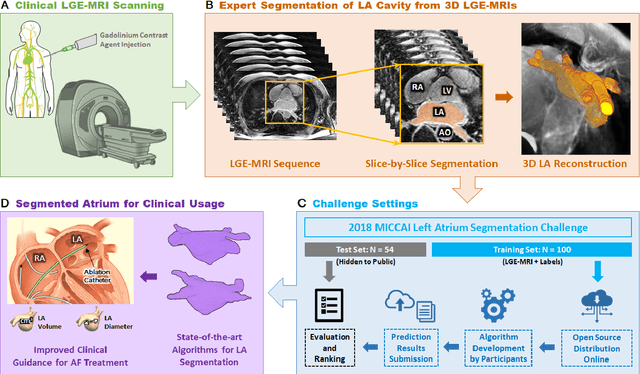
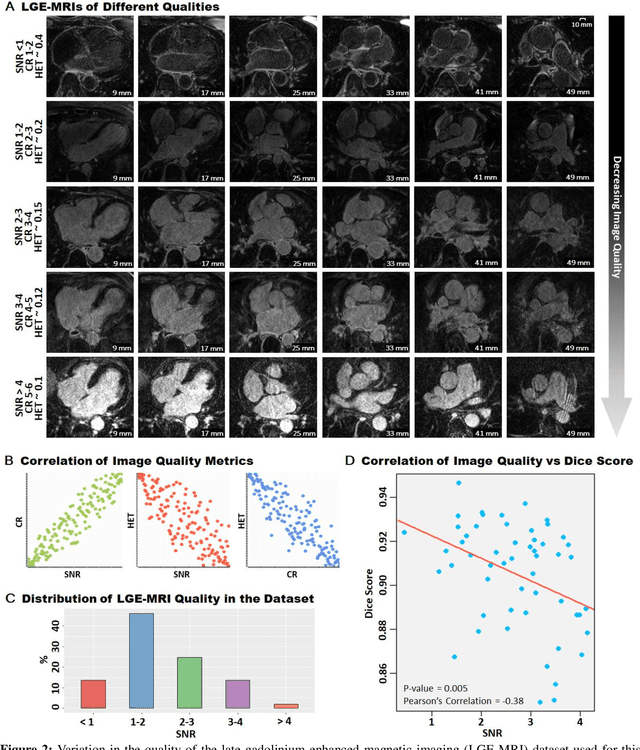
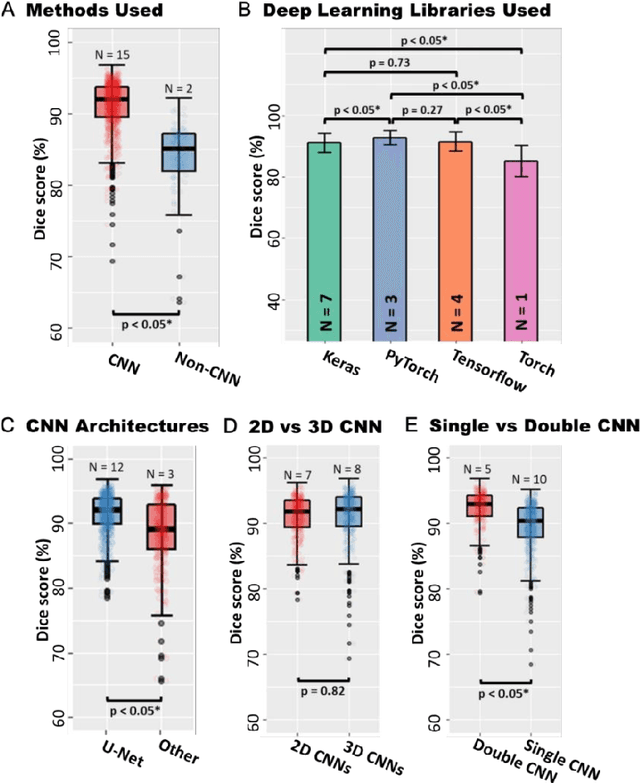
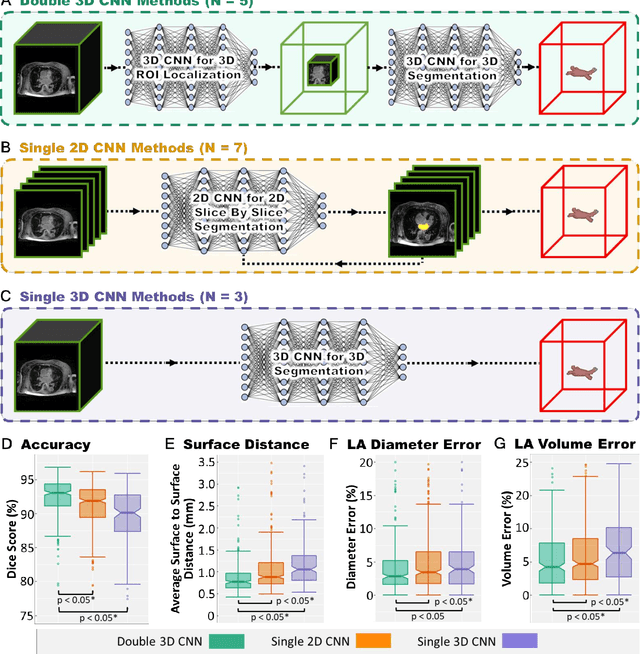
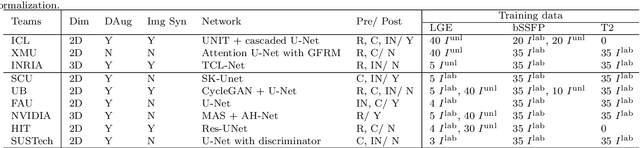
Abstract:Segmentation of cardiac images, particularly late gadolinium-enhanced magnetic resonance imaging (LGE-MRI) widely used for visualizing diseased cardiac structures, is a crucial first step for clinical diagnosis and treatment. However, direct segmentation of LGE-MRIs is challenging due to its attenuated contrast. Since most clinical studies have relied on manual and labor-intensive approaches, automatic methods are of high interest, particularly optimized machine learning approaches. To address this, we organized the "2018 Left Atrium Segmentation Challenge" using 154 3D LGE-MRIs, currently the world's largest cardiac LGE-MRI dataset, and associated labels of the left atrium segmented by three medical experts, ultimately attracting the participation of 27 international teams. In this paper, extensive analysis of the submitted algorithms using technical and biological metrics was performed by undergoing subgroup analysis and conducting hyper-parameter analysis, offering an overall picture of the major design choices of convolutional neural networks (CNNs) and practical considerations for achieving state-of-the-art left atrium segmentation. Results show the top method achieved a dice score of 93.2% and a mean surface to a surface distance of 0.7 mm, significantly outperforming prior state-of-the-art. Particularly, our analysis demonstrated that double, sequentially used CNNs, in which a first CNN is used for automatic region-of-interest localization and a subsequent CNN is used for refined regional segmentation, achieved far superior results than traditional methods and pipelines containing single CNNs. This large-scale benchmarking study makes a significant step towards much-improved segmentation methods for cardiac LGE-MRIs, and will serve as an important benchmark for evaluating and comparing the future works in the field.
Automatically Segmenting the Left Atrium from Cardiac Images Using Successive 3D U-Nets and a Contour Loss
Dec 06, 2018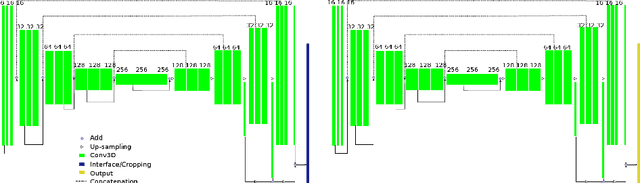
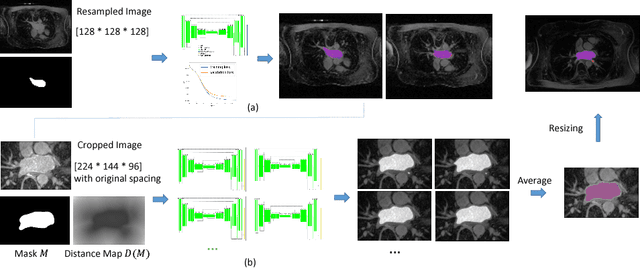
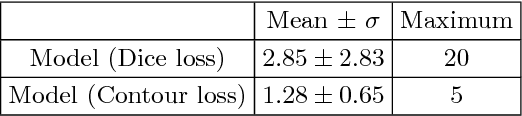
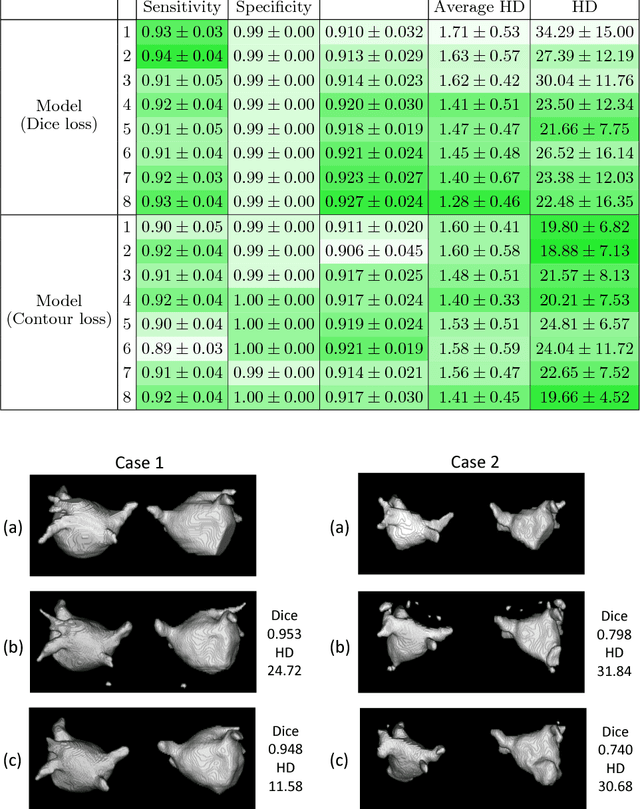
Abstract:Radiological imaging offers effective measurement of anatomy, which is useful in disease diagnosis and assessment. Previous study has shown that the left atrial wall remodeling can provide information to predict treatment outcome in atrial fibrillation. Nevertheless, the segmentation of the left atrial structures from medical images is still very time-consuming. Current advances in neural network may help creating automatic segmentation models that reduce the workload for clinicians. In this preliminary study, we propose automated, two-stage, three-dimensional U-Nets with convolutional neural network, for the challenging task of left atrial segmentation. Unlike previous two-dimensional image segmentation methods, we use 3D U-Nets to obtain the heart cavity directly in 3D. The dual 3D U-Net structure consists of, a first U-Net to coarsely segment and locate the left atrium, and a second U-Net to accurately segment the left atrium under higher resolution. In addition, we introduce a Contour loss based on additional distance information to adjust the final segmentation. We randomly split the data into training datasets (80 subjects) and validation datasets (20 subjects) to train multiple models, with different augmentation setting. Experiments show that the average Dice coefficients for validation datasets are around 0.91 - 0.92, the sensitivity around 0.90-0.94 and the specificity 0.99. Compared with traditional Dice loss, models trained with Contour loss in general offer smaller Hausdorff distance with similar Dice coefficient, and have less connected components in predictions. Finally, we integrate several trained models in an ensemble prediction to segment testing datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge