Buntheng Ly
EPIONE
Constraint-Based Model in Multimodal Learning to Improve Ventricular Arrhythmia Prediction
Dec 18, 2024Abstract:Cardiac disease evaluation depends on multiple diagnostic modalities: electrocardiogram (ECG) to diagnose abnormal heart rhythms, and imaging modalities such as Magnetic Resonance Imaging (MRI), Computed Tomography (CT) and echocardiography to detect signs of structural abnormalities. Each of these modalities brings complementary information for a better diagnosis of cardiac dysfunction. However, training a machine learning (ML) model with data from multiple modalities is a challenging task, as it increases the dimension space, while keeping constant the number of samples. In fact, as the dimension of the input space increases, the volume of data required for accurate generalisation grows exponentially. In this work, we address this issue, for the application of Ventricular Arrhythmia (VA) prediction, based on the combined clinical and CT imaging features, where we constrained the learning process on medical images (CT) based on the prior knowledge acquired from clinical data. The VA classifier is fed with features extracted from a 3D myocardium thickness map (TM) of the left ventricle. The TM is generated by our pipeline from the imaging input and a Graph Convolutional Network is used as the feature extractor of the 3D TM. We introduce a novel Sequential Fusion method and evaluate its performance against traditional Early Fusion techniques and single-modality models. The crossvalidation results show that the Sequential Fusion model achieved the highest average scores of 80.7% $\pm$ 4.4 Sensitivity and 73.1% $\pm$ 6.0 F1 score, outperforming the Early Fusion model at 65.0% $\pm$ 8.9 Sensitivity and 63.1% $\pm$ 6.3 F1 score. Both fusion models achieved better scores than the single-modality models, where the average Sensitivity and F1 score are 62.8% $\pm$ 10.1; 52.1% $\pm$ 6.5 for the clinical data modality and 62.9% $\pm$ 6.3; 60.7% $\pm$ 5.3 for the medical images modality.
CrossMoDA 2021 challenge: Benchmark of Cross-Modality Domain Adaptation techniques for Vestibular Schwnannoma and Cochlea Segmentation
Jan 08, 2022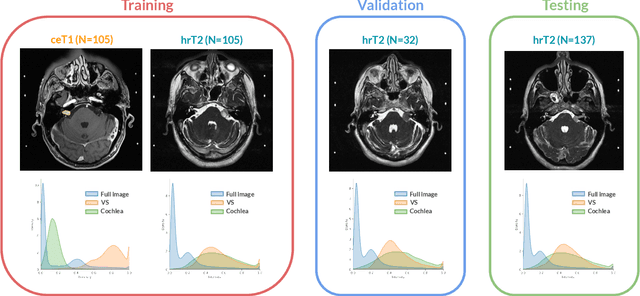
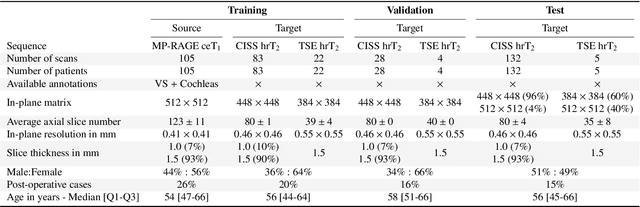
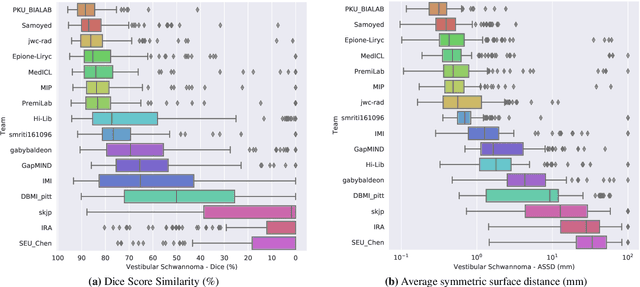
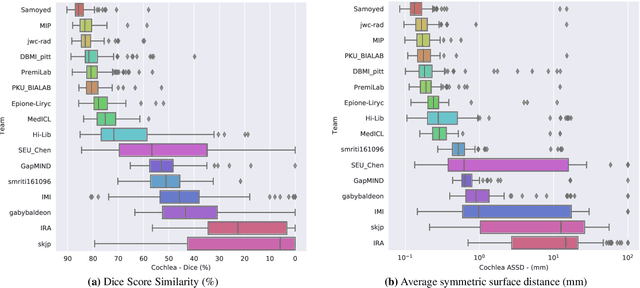
Abstract:Domain Adaptation (DA) has recently raised strong interests in the medical imaging community. While a large variety of DA techniques has been proposed for image segmentation, most of these techniques have been validated either on private datasets or on small publicly available datasets. Moreover, these datasets mostly addressed single-class problems. To tackle these limitations, the Cross-Modality Domain Adaptation (crossMoDA) challenge was organised in conjunction with the 24th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2021). CrossMoDA is the first large and multi-class benchmark for unsupervised cross-modality DA. The challenge's goal is to segment two key brain structures involved in the follow-up and treatment planning of vestibular schwannoma (VS): the VS and the cochleas. Currently, the diagnosis and surveillance in patients with VS are performed using contrast-enhanced T1 (ceT1) MRI. However, there is growing interest in using non-contrast sequences such as high-resolution T2 (hrT2) MRI. Therefore, we created an unsupervised cross-modality segmentation benchmark. The training set provides annotated ceT1 (N=105) and unpaired non-annotated hrT2 (N=105). The aim was to automatically perform unilateral VS and bilateral cochlea segmentation on hrT2 as provided in the testing set (N=137). A total of 16 teams submitted their algorithm for the evaluation phase. The level of performance reached by the top-performing teams is strikingly high (best median Dice - VS:88.4%; Cochleas:85.7%) and close to full supervision (median Dice - VS:92.5%; Cochleas:87.7%). All top-performing methods made use of an image-to-image translation approach to transform the source-domain images into pseudo-target-domain images. A segmentation network was then trained using these generated images and the manual annotations provided for the source image.
Cardiac Segmentation on Late Gadolinium Enhancement MRI: A Benchmark Study from Multi-Sequence Cardiac MR Segmentation Challenge
Jun 22, 2020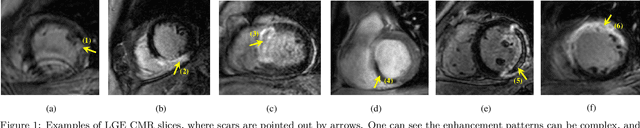
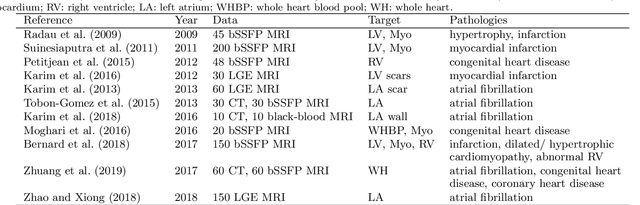
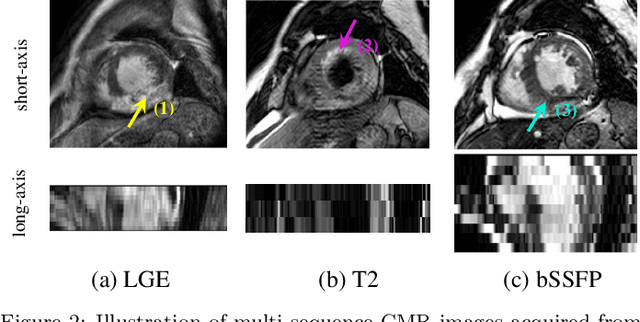
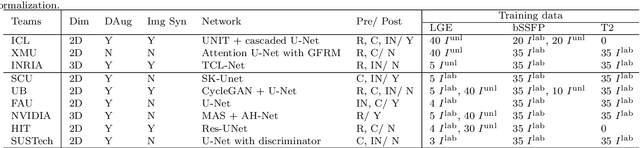
Abstract:Accurate computing, analysis and modeling of the ventricles and myocardium from medical images are important, especially in the diagnosis and treatment management for patients suffering from myocardial infarction (MI). Late gadolinium enhancement (LGE) cardiac magnetic resonance (CMR) provides an important protocol to visualize MI. However, automated segmentation of LGE CMR is still challenging, due to the indistinguishable boundaries, heterogeneous intensity distribution and complex enhancement patterns of pathological myocardium from LGE CMR. Furthermore, compared with the other sequences LGE CMR images with gold standard labels are particularly limited, which represents another obstacle for developing novel algorithms for automatic segmentation of LGE CMR. This paper presents the selective results from the Multi-Sequence Cardiac MR (MS-CMR) Segmentation challenge, in conjunction with MICCAI 2019. The challenge offered a data set of paired MS-CMR images, including auxiliary CMR sequences as well as LGE CMR, from 45 patients who underwent cardiomyopathy. It was aimed to develop new algorithms, as well as benchmark existing ones for LGE CMR segmentation and compare them objectively. In addition, the paired MS-CMR images could enable algorithms to combine the complementary information from the other sequences for the segmentation of LGE CMR. Nine representative works were selected for evaluation and comparisons, among which three methods are unsupervised methods and the other six are supervised. The results showed that the average performance of the nine methods was comparable to the inter-observer variations. The success of these methods was mainly attributed to the inclusion of the auxiliary sequences from the MS-CMR images, which provide important label information for the training of deep neural networks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge