Marc Pàmies
Salamandra Technical Report
Feb 12, 2025Abstract:This work introduces Salamandra, a suite of open-source decoder-only large language models available in three different sizes: 2, 7, and 40 billion parameters. The models were trained from scratch on highly multilingual data that comprises text in 35 European languages and code. Our carefully curated corpus is made exclusively from open-access data compiled from a wide variety of sources. Along with the base models, supplementary checkpoints that were fine-tuned on public-domain instruction data are also released for chat applications. Additionally, we also share our preliminary experiments on multimodality, which serve as proof-of-concept to showcase potential applications for the Salamandra family. Our extensive evaluations on multilingual benchmarks reveal that Salamandra has strong capabilities, achieving competitive performance when compared to similarly sized open-source models. We provide comprehensive evaluation results both on standard downstream tasks as well as key aspects related to bias and safety.With this technical report, we intend to promote open science by sharing all the details behind our design choices, data curation strategy and evaluation methodology. In addition to that, we deviate from the usual practice by making our training and evaluation scripts publicly accessible. We release all models under a permissive Apache 2.0 license in order to foster future research and facilitate commercial use, thereby contributing to the open-source ecosystem of large language models.
BLOOM: A 176B-Parameter Open-Access Multilingual Language Model
Nov 09, 2022Abstract:Large language models (LLMs) have been shown to be able to perform new tasks based on a few demonstrations or natural language instructions. While these capabilities have led to widespread adoption, most LLMs are developed by resource-rich organizations and are frequently kept from the public. As a step towards democratizing this powerful technology, we present BLOOM, a 176B-parameter open-access language model designed and built thanks to a collaboration of hundreds of researchers. BLOOM is a decoder-only Transformer language model that was trained on the ROOTS corpus, a dataset comprising hundreds of sources in 46 natural and 13 programming languages (59 in total). We find that BLOOM achieves competitive performance on a wide variety of benchmarks, with stronger results after undergoing multitask prompted finetuning. To facilitate future research and applications using LLMs, we publicly release our models and code under the Responsible AI License.
BigBIO: A Framework for Data-Centric Biomedical Natural Language Processing
Jun 30, 2022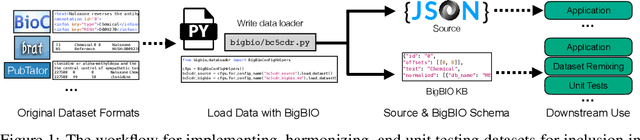
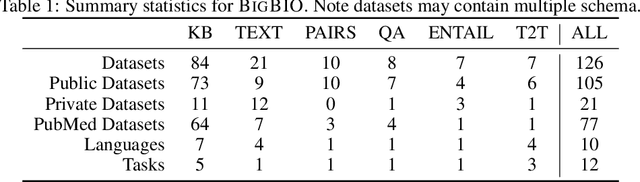
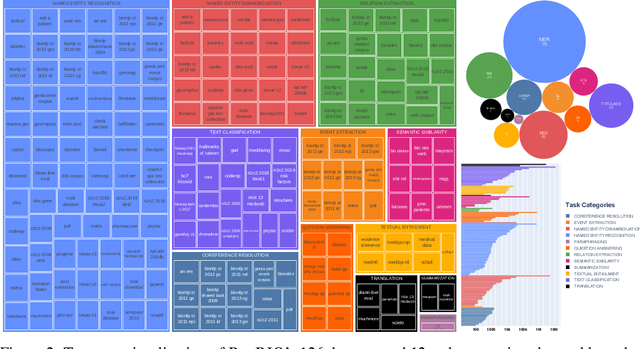
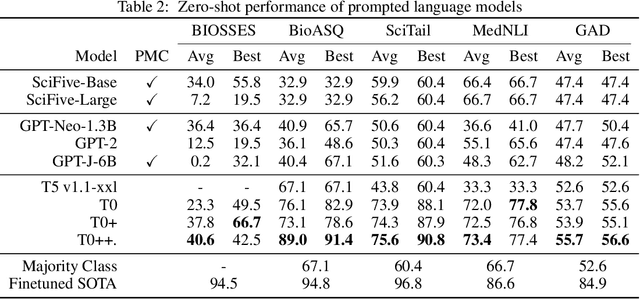
Abstract:Training and evaluating language models increasingly requires the construction of meta-datasets --diverse collections of curated data with clear provenance. Natural language prompting has recently lead to improved zero-shot generalization by transforming existing, supervised datasets into a diversity of novel pretraining tasks, highlighting the benefits of meta-dataset curation. While successful in general-domain text, translating these data-centric approaches to biomedical language modeling remains challenging, as labeled biomedical datasets are significantly underrepresented in popular data hubs. To address this challenge, we introduce BigBIO a community library of 126+ biomedical NLP datasets, currently covering 12 task categories and 10+ languages. BigBIO facilitates reproducible meta-dataset curation via programmatic access to datasets and their metadata, and is compatible with current platforms for prompt engineering and end-to-end few/zero shot language model evaluation. We discuss our process for task schema harmonization, data auditing, contribution guidelines, and outline two illustrative use cases: zero-shot evaluation of biomedical prompts and large-scale, multi-task learning. BigBIO is an ongoing community effort and is available at https://github.com/bigscience-workshop/biomedical
Biomedical and Clinical Language Models for Spanish: On the Benefits of Domain-Specific Pretraining in a Mid-Resource Scenario
Sep 17, 2021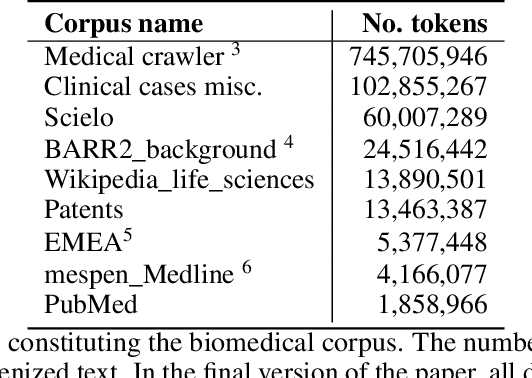
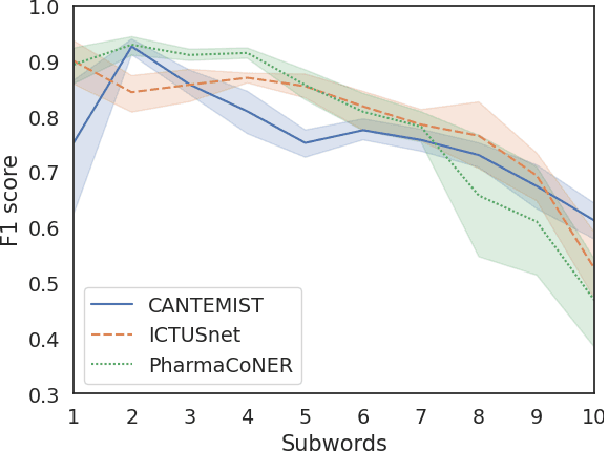
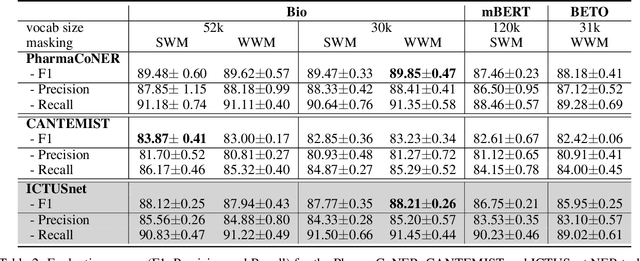
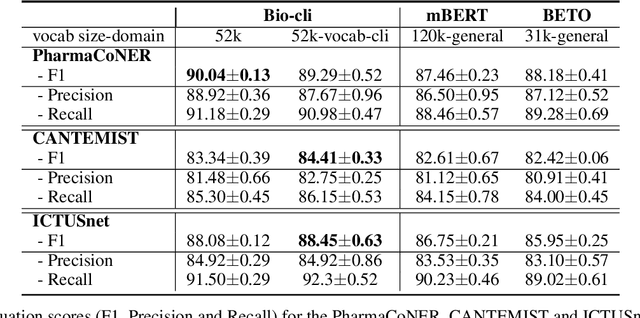
Abstract:This work presents biomedical and clinical language models for Spanish by experimenting with different pretraining choices, such as masking at word and subword level, varying the vocabulary size and testing with domain data, looking for better language representations. Interestingly, in the absence of enough clinical data to train a model from scratch, we applied mixed-domain pretraining and cross-domain transfer approaches to generate a performant bio-clinical model suitable for real-world clinical data. We evaluated our models on Named Entity Recognition (NER) tasks for biomedical documents and challenging hospital discharge reports. When compared against the competitive mBERT and BETO models, we outperform them in all NER tasks by a significant margin. Finally, we studied the impact of the model's vocabulary on the NER performances by offering an interesting vocabulary-centric analysis. The results confirm that domain-specific pretraining is fundamental to achieving higher performances in downstream NER tasks, even within a mid-resource scenario. To the best of our knowledge, we provide the first biomedical and clinical transformer-based pretrained language models for Spanish, intending to boost native Spanish NLP applications in biomedicine. Our best models are freely available in the HuggingFace hub: https://huggingface.co/BSC-TeMU.
Spanish Language Models
Aug 13, 2021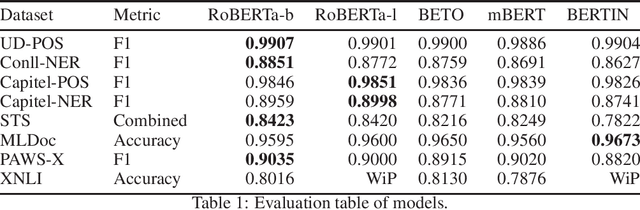
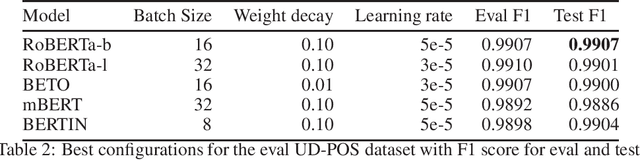
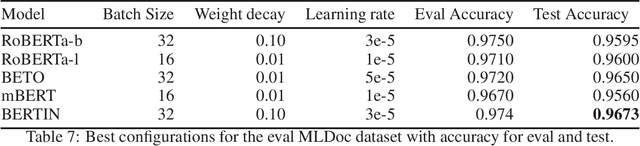
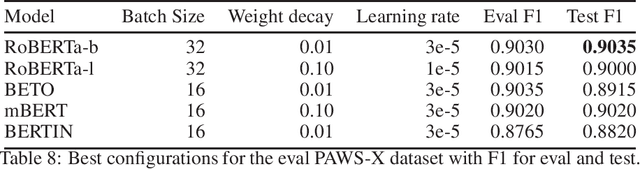
Abstract:This paper presents the Spanish RoBERTa-base and RoBERTa-large models, as well as the corresponding performance evaluations. Both models were pre-trained using the largest Spanish corpus known to date, with a total of 570GB of clean and deduplicated text processed for this work, compiled from the web crawlings performed by the National Library of Spain from 2009 to 2019. We extended the current evaluation datasets with an extractive Question Answering dataset and our models outperform the existing Spanish models across tasks and settings.
XED: A Multilingual Dataset for Sentiment Analysis and Emotion Detection
Nov 06, 2020



Abstract:We introduce XED, a multilingual fine-grained emotion dataset. The dataset consists of human-annotated Finnish (25k) and English sentences (30k), as well as projected annotations for 30 additional languages, providing new resources for many low-resource languages. We use Plutchik's core emotions to annotate the dataset with the addition of neutral to create a multilabel multiclass dataset. The dataset is carefully evaluated using language-specific BERT models and SVMs to show that XED performs on par with other similar datasets and is therefore a useful tool for sentiment analysis and emotion detection.
LT@Helsinki at SemEval-2020 Task 12: Multilingual or language-specific BERT?
Aug 03, 2020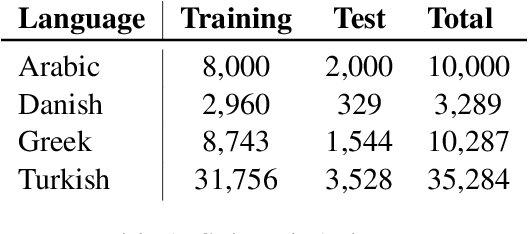

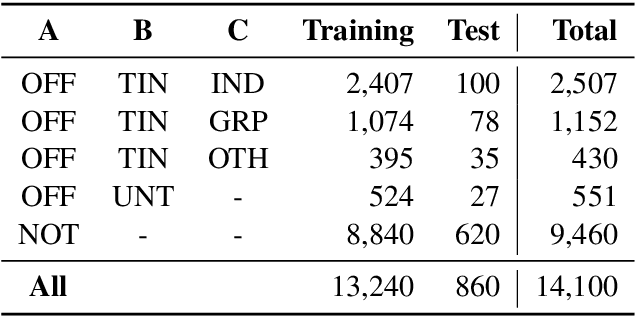

Abstract:This paper presents the different models submitted by the LT@Helsinki team for the SemEval 2020 Shared Task 12. Our team participated in sub-tasks A and C; titled offensive language identification and offense target identification, respectively. In both cases we used the so-called Bidirectional Encoder Representation from Transformer (BERT), a model pre-trained by Google and fine-tuned by us on the OLID and SOLID datasets. The results show that offensive tweet classification is one of several language-based tasks where BERT can achieve state-of-the-art results.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge