Marta Villegas
XDoGE: Multilingual Data Reweighting to Enhance Language Inclusivity in LLMs
Dec 11, 2025



Abstract:Current large language models (LLMs) are trained on massive amounts of text data, primarily from a few dominant languages. Studies suggest that this over-reliance on high-resource languages, such as English, hampers LLM performance in mid- and low-resource languages. To mitigate this problem, we propose to (i) optimize the language distribution by training a small proxy model within a domain-reweighing DoGE algorithm that we extend to XDoGE for a multilingual setup, and (ii) rescale the data and train a full-size model with the established language weights either from scratch or within a continual pre-training phase (CPT). We target six languages possessing a variety of geographic and intra- and inter-language-family relations, namely, English and Spanish (high-resource), Portuguese and Catalan (mid-resource), Galician and Basque (low-resource). We experiment with Salamandra-2b, which is a promising model for these languages. We investigate the effects of substantial data repetition on minor languages and under-sampling on dominant languages using the IberoBench framework for quantitative evaluation. Finally, we release a new promising IberianLLM-7B-Instruct model centering on Iberian languages and English that we pretrained from scratch and further improved using CPT with the XDoGE weights.
ACADATA: Parallel Dataset of Academic Data for Machine Translation
Oct 14, 2025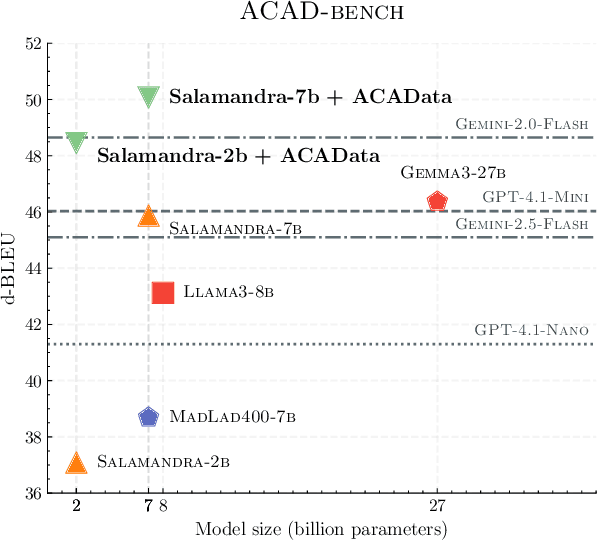
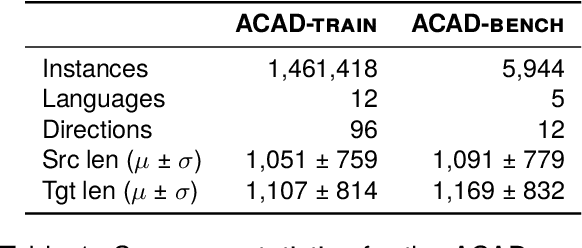
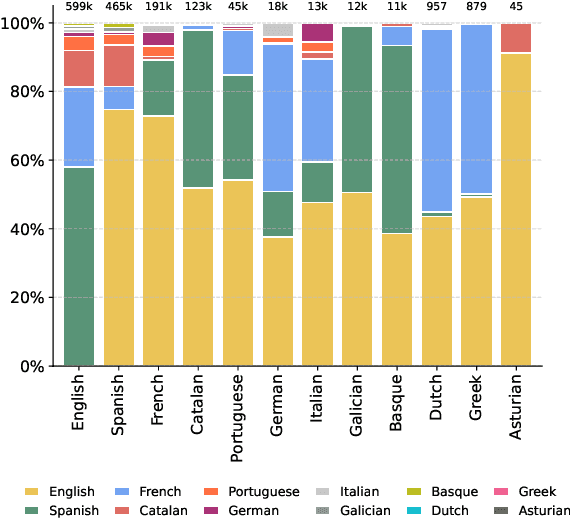
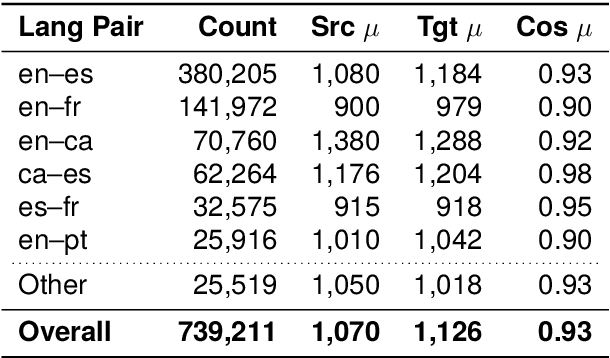
Abstract:We present ACADATA, a high-quality parallel dataset for academic translation, that consists of two subsets: ACAD-TRAIN, which contains approximately 1.5 million author-generated paragraph pairs across 96 language directions and ACAD-BENCH, a curated evaluation set of almost 6,000 translations covering 12 directions. To validate its utility, we fine-tune two Large Language Models (LLMs) on ACAD-TRAIN and benchmark them on ACAD-BENCH against specialized machine-translation systems, general-purpose, open-weight LLMs, and several large-scale proprietary models. Experimental results demonstrate that fine-tuning on ACAD-TRAIN leads to improvements in academic translation quality by +6.1 and +12.4 d-BLEU points on average for 7B and 2B models respectively, while also improving long-context translation in a general domain by up to 24.9% when translating out of English. The fine-tuned top-performing model surpasses the best propietary and open-weight models on academic translation domain. By releasing ACAD-TRAIN, ACAD-BENCH and the fine-tuned models, we provide the community with a valuable resource to advance research in academic domain and long-context translation.
Breaking Language Barriers in Visual Language Models via Multilingual Textual Regularization
Mar 28, 2025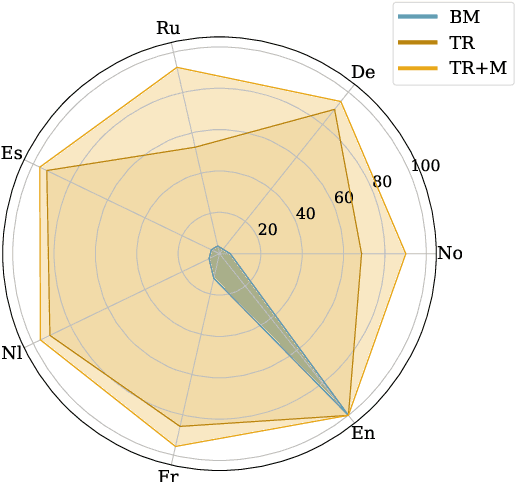
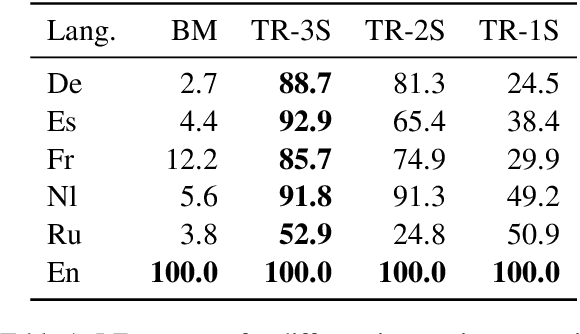
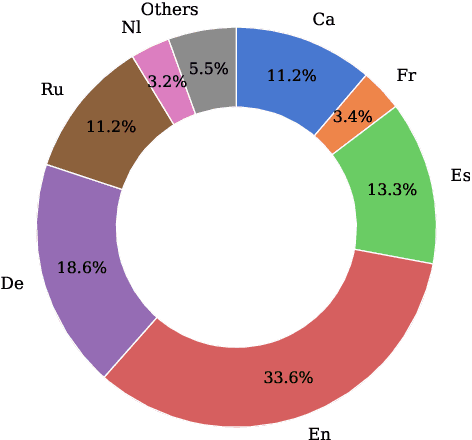
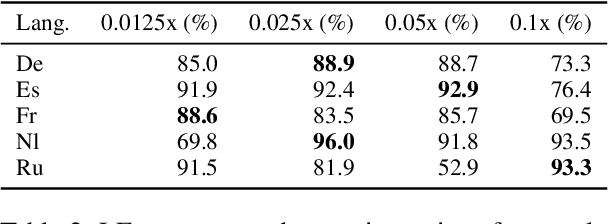
Abstract:Rapid advancements in Visual Language Models (VLMs) have transformed multimodal understanding but are often constrained by generating English responses regardless of the input language. This phenomenon has been termed as Image-induced Fidelity Loss (IFL) and stems from limited multimodal multilingual training data. To address this, we propose a continuous multilingual integration strategy that injects text-only multilingual data during visual instruction tuning, preserving the language model's original multilingual capabilities. Extensive evaluations demonstrate that our approach significantly improves linguistic fidelity across languages without degradation in visual performance. We also explore model merging, which improves language fidelity but comes at the cost of visual performance. In contrast, our core method achieves robust multilingual alignment without trade-offs, offering a scalable and effective path to mitigating IFL for global VLM adoption.
Salamandra Technical Report
Feb 12, 2025Abstract:This work introduces Salamandra, a suite of open-source decoder-only large language models available in three different sizes: 2, 7, and 40 billion parameters. The models were trained from scratch on highly multilingual data that comprises text in 35 European languages and code. Our carefully curated corpus is made exclusively from open-access data compiled from a wide variety of sources. Along with the base models, supplementary checkpoints that were fine-tuned on public-domain instruction data are also released for chat applications. Additionally, we also share our preliminary experiments on multimodality, which serve as proof-of-concept to showcase potential applications for the Salamandra family. Our extensive evaluations on multilingual benchmarks reveal that Salamandra has strong capabilities, achieving competitive performance when compared to similarly sized open-source models. We provide comprehensive evaluation results both on standard downstream tasks as well as key aspects related to bias and safety.With this technical report, we intend to promote open science by sharing all the details behind our design choices, data curation strategy and evaluation methodology. In addition to that, we deviate from the usual practice by making our training and evaluation scripts publicly accessible. We release all models under a permissive Apache 2.0 license in order to foster future research and facilitate commercial use, thereby contributing to the open-source ecosystem of large language models.
Mass-Editing Memory with Attention in Transformers: A cross-lingual exploration of knowledge
Feb 04, 2025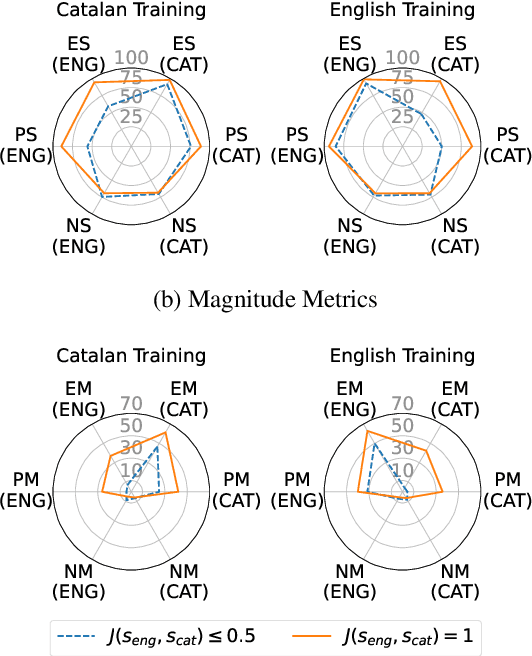
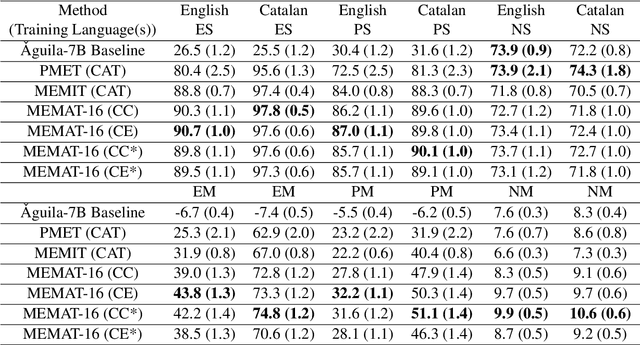
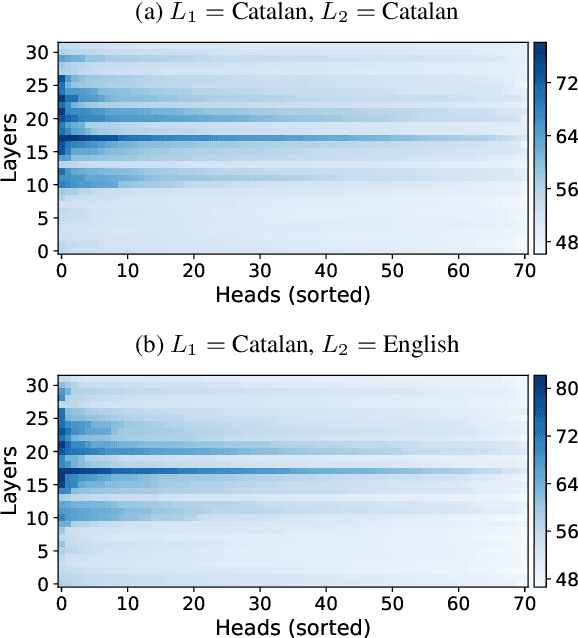
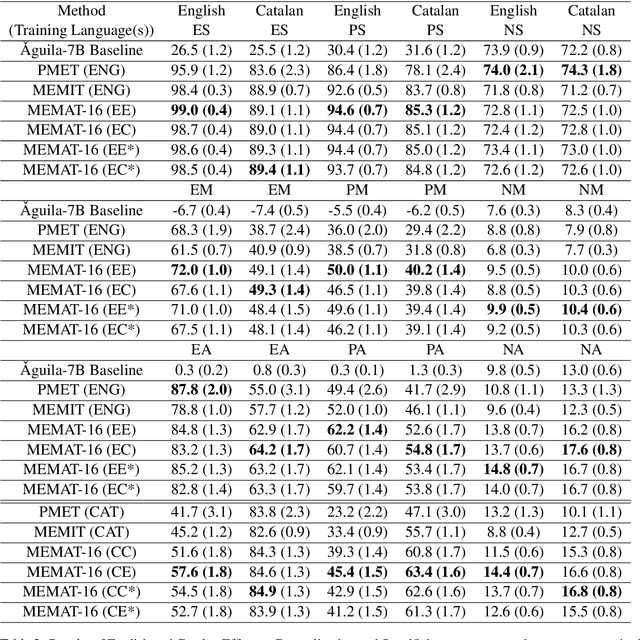
Abstract:Recent research has explored methods for updating and modifying factual knowledge in large language models, often focusing on specific multi-layer perceptron blocks. This study expands on this work by examining the effectiveness of existing knowledge editing methods across languages and delving into the role of attention mechanisms in this process. Drawing from the insights gained, we propose Mass-Editing Memory with Attention in Transformers (MEMAT), a method that achieves significant improvements in all metrics while requiring minimal parameter modifications. MEMAT delivers a remarkable 10% increase in magnitude metrics, benefits languages not included in the training data and also demonstrates a high degree of portability. Our code and data are at https://github.com/dtamayo-nlp/MEMAT.
The Catalan Language CLUB
Dec 03, 2021

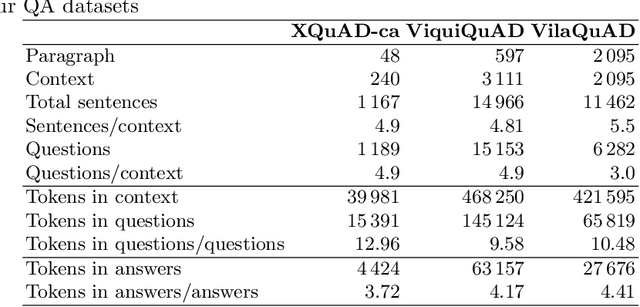
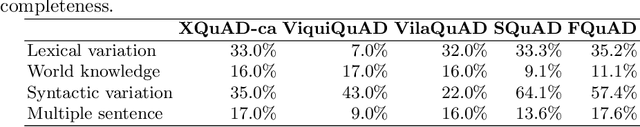
Abstract:The Catalan Language Understanding Benchmark (CLUB) encompasses various datasets representative of different NLU tasks that enable accurate evaluations of language models, following the General Language Understanding Evaluation (GLUE) example. It is part of AINA and PlanTL, two public funding initiatives to empower the Catalan language in the Artificial Intelligence era.
Spanish Legalese Language Model and Corpora
Oct 23, 2021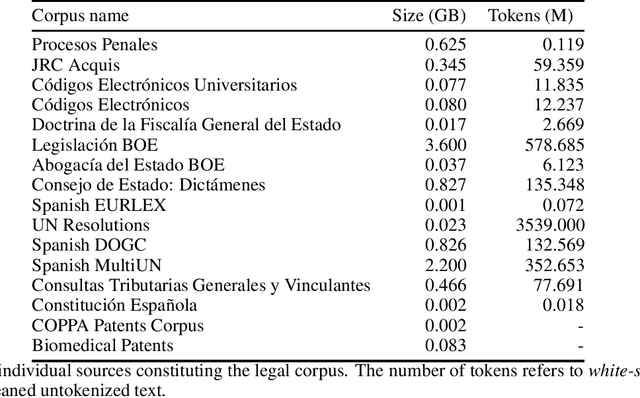
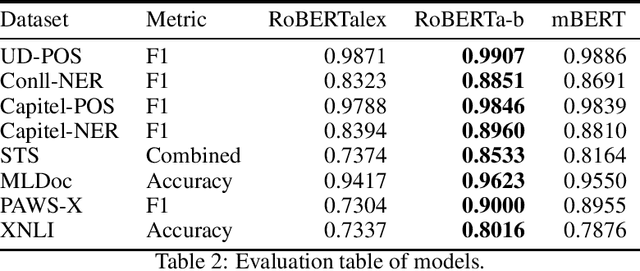
Abstract:There are many Language Models for the English language according to its worldwide relevance. However, for the Spanish language, even if it is a widely spoken language, there are very few Spanish Language Models which result to be small and too general. Legal slang could be think of a Spanish variant on its own as it is very complicated in vocabulary, semantics and phrase understanding. For this work we gathered legal-domain corpora from different sources, generated a model and evaluated against Spanish general domain tasks. The model provides reasonable results in those tasks.
Biomedical and Clinical Language Models for Spanish: On the Benefits of Domain-Specific Pretraining in a Mid-Resource Scenario
Sep 17, 2021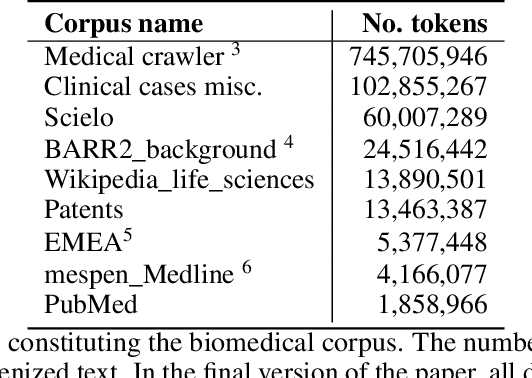
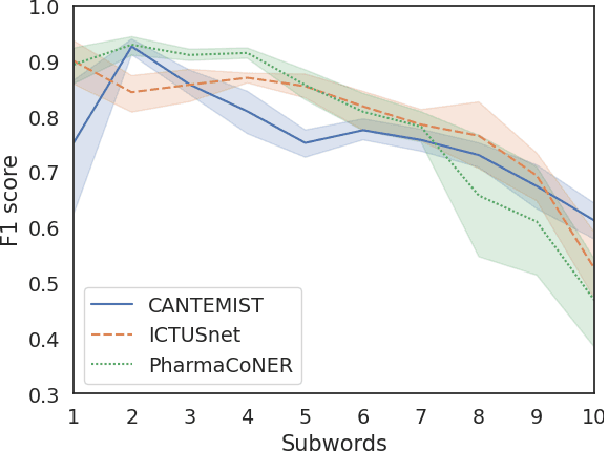
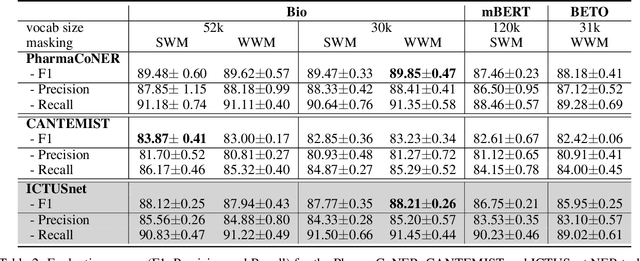
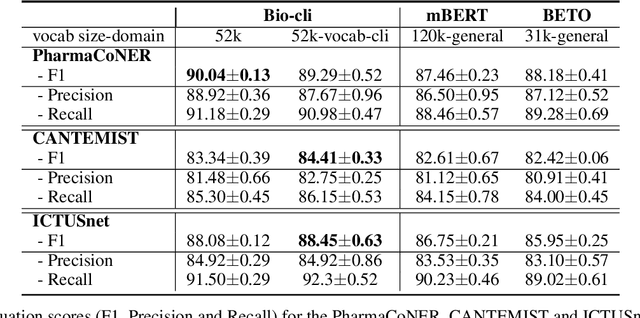
Abstract:This work presents biomedical and clinical language models for Spanish by experimenting with different pretraining choices, such as masking at word and subword level, varying the vocabulary size and testing with domain data, looking for better language representations. Interestingly, in the absence of enough clinical data to train a model from scratch, we applied mixed-domain pretraining and cross-domain transfer approaches to generate a performant bio-clinical model suitable for real-world clinical data. We evaluated our models on Named Entity Recognition (NER) tasks for biomedical documents and challenging hospital discharge reports. When compared against the competitive mBERT and BETO models, we outperform them in all NER tasks by a significant margin. Finally, we studied the impact of the model's vocabulary on the NER performances by offering an interesting vocabulary-centric analysis. The results confirm that domain-specific pretraining is fundamental to achieving higher performances in downstream NER tasks, even within a mid-resource scenario. To the best of our knowledge, we provide the first biomedical and clinical transformer-based pretrained language models for Spanish, intending to boost native Spanish NLP applications in biomedicine. Our best models are freely available in the HuggingFace hub: https://huggingface.co/BSC-TeMU.
Spanish Biomedical Crawled Corpus: A Large, Diverse Dataset for Spanish Biomedical Language Models
Sep 16, 2021
Abstract:We introduce CoWeSe (the Corpus Web Salud Espa\~nol), the largest Spanish biomedical corpus to date, consisting of 4.5GB (about 750M tokens) of clean plain text. CoWeSe is the result of a massive crawler on 3000 Spanish domains executed in 2020. The corpus is openly available and already preprocessed. CoWeSe is an important resource for biomedical and health NLP in Spanish and has already been employed to train domain-specific language models and to produce word embbedings. We released the CoWeSe corpus under a Creative Commons Attribution 4.0 International license, both in Zenodo (\url{https://zenodo.org/record/4561971\#.YTI5SnVKiEA}).
Spanish Language Models
Aug 13, 2021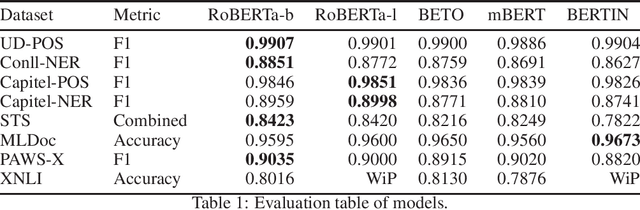
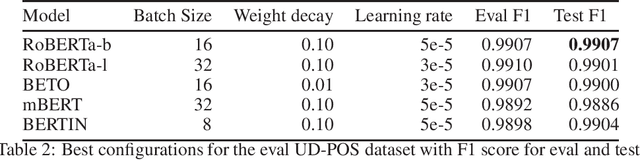
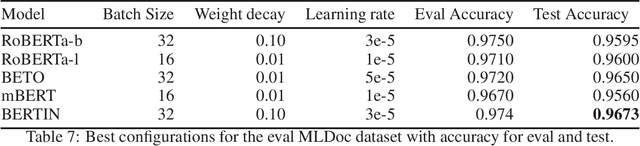
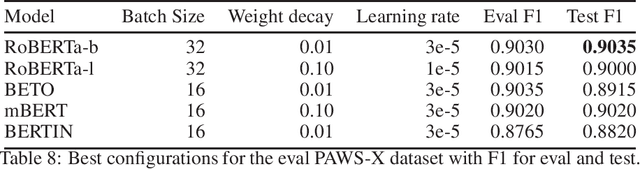
Abstract:This paper presents the Spanish RoBERTa-base and RoBERTa-large models, as well as the corresponding performance evaluations. Both models were pre-trained using the largest Spanish corpus known to date, with a total of 570GB of clean and deduplicated text processed for this work, compiled from the web crawlings performed by the National Library of Spain from 2009 to 2019. We extended the current evaluation datasets with an extractive Question Answering dataset and our models outperform the existing Spanish models across tasks and settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge