Konstantinos Bougiatiotis
From Primes to Paths: Enabling Fast Multi-Relational Graph Analysis
Nov 17, 2024Abstract:Multi-relational networks capture intricate relationships in data and have diverse applications across fields such as biomedical, financial, and social sciences. As networks derived from increasingly large datasets become more common, identifying efficient methods for representing and analyzing them becomes crucial. This work extends the Prime Adjacency Matrices (PAMs) framework, which employs prime numbers to represent distinct relations within a network uniquely. This enables a compact representation of a complete multi-relational graph using a single adjacency matrix, which, in turn, facilitates quick computation of multi-hop adjacency matrices. In this work, we enhance the framework by introducing a lossless algorithm for calculating the multi-hop matrices and propose the Bag of Paths (BoP) representation, a versatile feature extraction methodology for various graph analytics tasks, at the node, edge, and graph level. We demonstrate the efficiency of the framework across various tasks and datasets, showing that simple BoP-based models perform comparably to or better than commonly used neural models while offering improved speed and interpretability.
Financial misstatement detection: a realistic evaluation
May 27, 2023


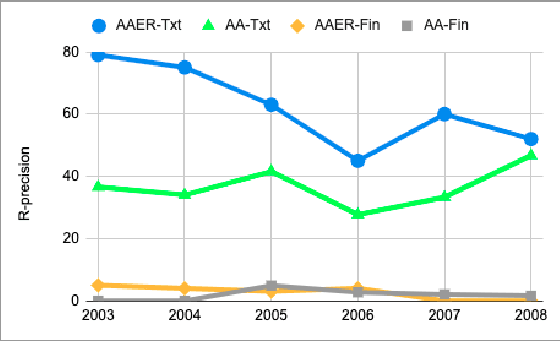
Abstract:In this work, we examine the evaluation process for the task of detecting financial reports with a high risk of containing a misstatement. This task is often referred to, in the literature, as ``misstatement detection in financial reports''. We provide an extensive review of the related literature. We propose a new, realistic evaluation framework for the task which, unlike a large part of the previous work: (a) focuses on the misstatement class and its rarity, (b) considers the dimension of time when splitting data into training and test and (c) considers the fact that misstatements can take a long time to detect. Most importantly, we show that the evaluation process significantly affects system performance, and we analyze the performance of different models and feature types in the new realistic framework.
* 9 pages, ICAIF2021
Analysing Biomedical Knowledge Graphs using Prime Adjacency Matrices
May 17, 2023



Abstract:Most phenomena related to biomedical tasks are inherently complex, and in many cases, are expressed as signals on biomedical Knowledge Graphs (KGs). In this work, we introduce the use of a new representation framework, the Prime Adjacency Matrix (PAM) for biomedical KGs, which allows for very efficient network analysis. PAM utilizes prime numbers to enable representing the whole KG with a single adjacency matrix and the fast computation of multiple properties of the network. We illustrate the applicability of the framework in the biomedical domain by working on different biomedical knowledge graphs and by providing two case studies: one on drug-repurposing for COVID-19 and one on important metapath extraction. We show that we achieve better results than the original proposed workflows, using very simple methods that require no training, in considerably less time.
Efficient multi-relational network representation using primes
Sep 14, 2022



Abstract:Multi-relational networks play an important role in today's world and are utilized to capture complex relationships between the data. Their applications span many domains such as biomedical, financial, social, etc., and because of their increasing usability, it becomes crucial to find efficient ways to deal with the added complexity of multiple layers. In this work, we propose a novel approach to represent these complex networks using a single aggregated adjacency matrix, by utilizing primes as surrogates for the relations. Due to the fundamental theorem of arithmetic, this allows for a lossless, compact representation of the whole multi-relational graph, using a single adjacency matrix. Moreover, this representation enables the fast computation of multi-hop adjacency matrices, that can be useful for a variety of downstream tasks. We present simple and complex tasks in which this representation can be useful and showcase its efficiency and performance. Finally, we also provide insights on the advantages and the open challenges that still need to be addressed and motivate future work.
DICoE@FinSim-3: Financial Hypernym Detection using Augmented Terms and Distance-based Features
Sep 30, 2021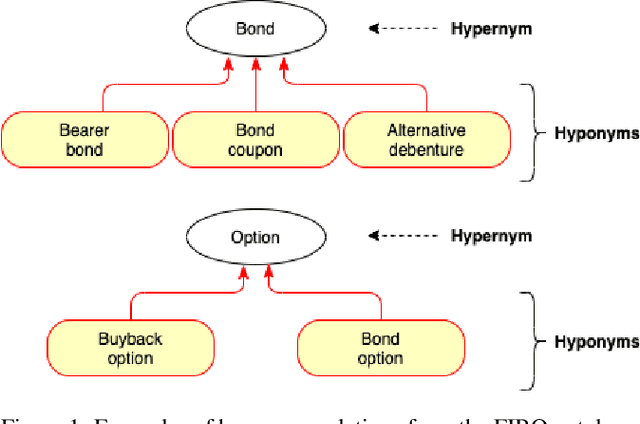
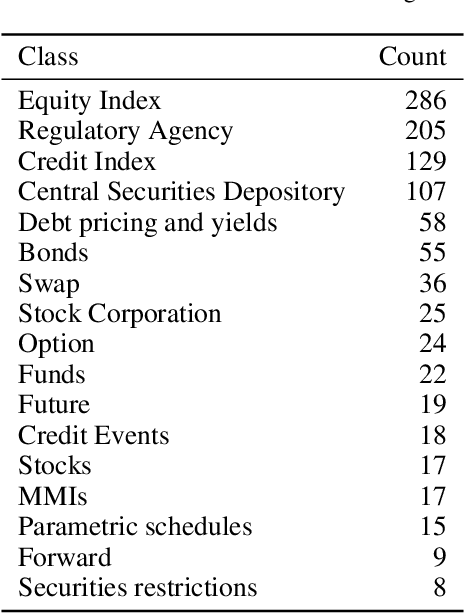
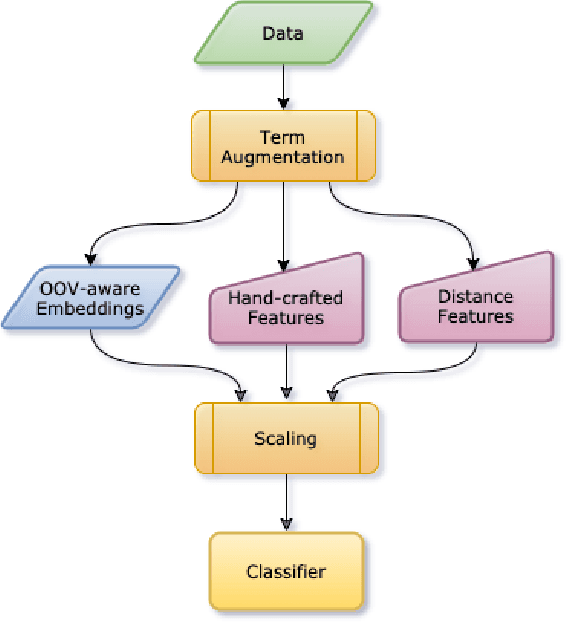
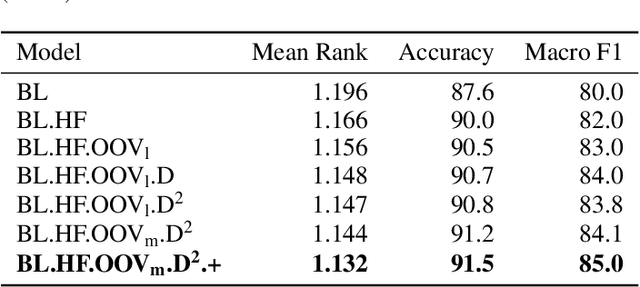
Abstract:We present the submission of team DICoE for FinSim-3, the 3rd Shared Task on Learning Semantic Similarities for the Financial Domain. The task provides a set of terms in the financial domain and requires to classify them into the most relevant hypernym from a financial ontology. After augmenting the terms with their Investopedia definitions, our system employs a Logistic Regression classifier over financial word embeddings and a mix of hand-crafted and distance-based features. Also, for the first time in this task, we employ different replacement methods for out-of-vocabulary terms, leading to improved performance. Finally, we have also experimented with word representations generated from various financial corpora. Our best-performing submission ranked 4th on the task's leaderboard.
Overview of BioASQ 2020: The eighth BioASQ challenge on Large-Scale Biomedical Semantic Indexing and Question Answering
Jun 28, 2021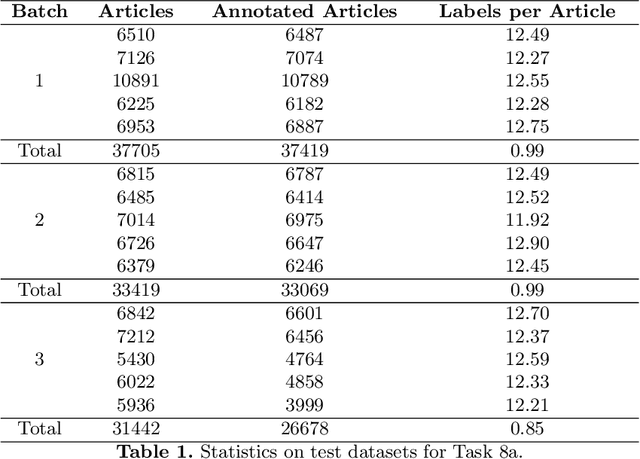
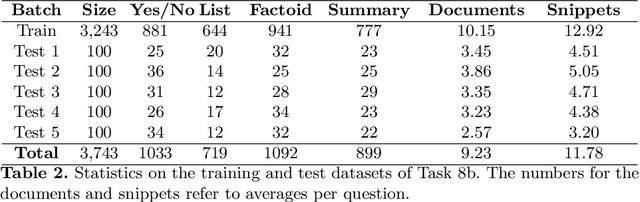
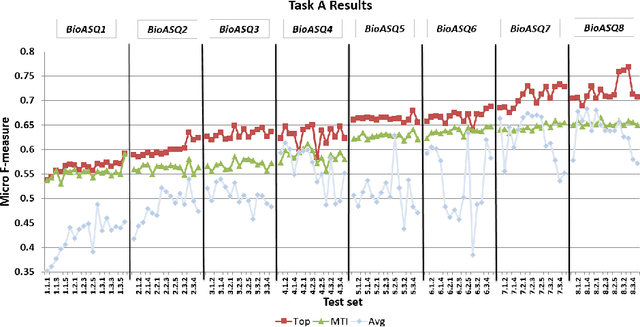
Abstract:In this paper, we present an overview of the eighth edition of the BioASQ challenge, which ran as a lab in the Conference and Labs of the Evaluation Forum (CLEF) 2020. BioASQ is a series of challenges aiming at the promotion of systems and methodologies for large-scale biomedical semantic indexing and question answering. To this end, shared tasks are organized yearly since 2012, where different teams develop systems that compete on the same demanding benchmark datasets that represent the real information needs of experts in the biomedical domain. This year, the challenge has been extended with the introduction of a new task on medical semantic indexing in Spanish. In total, 34 teams with more than 100 systems participated in the three tasks of the challenge. As in previous years, the results of the evaluation reveal that the top-performing systems managed to outperform the strong baselines, which suggests that state-of-the-art systems keep pushing the frontier of research through continuous improvements.
* 21 pages, 10 tables, 3 figures
Results of the seventh edition of the BioASQ Challenge
Jun 16, 2020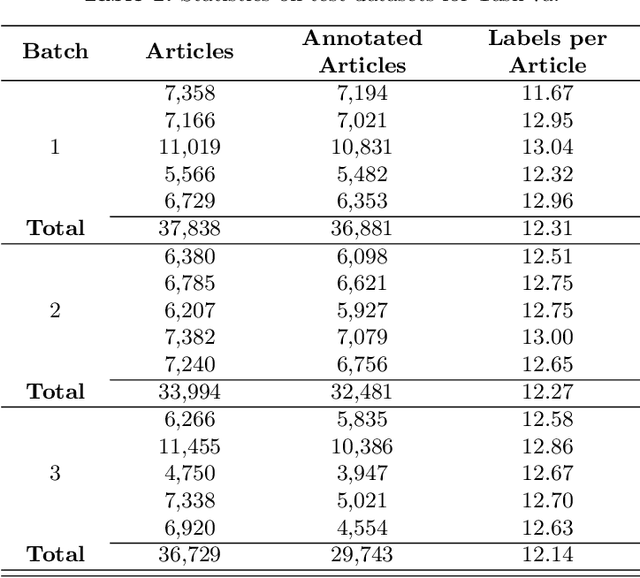
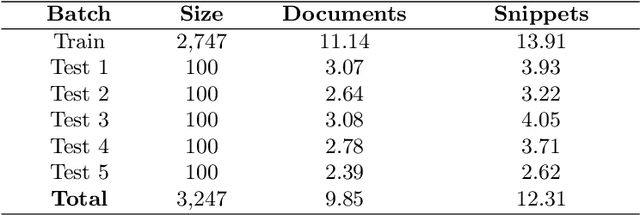
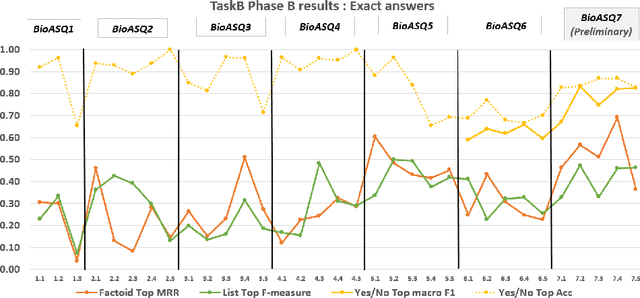
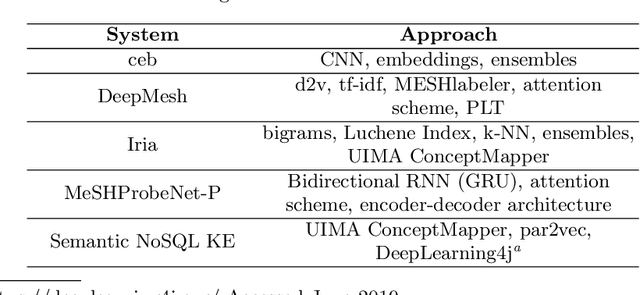
Abstract:The results of the seventh edition of the BioASQ challenge are presented in this paper. The aim of the BioASQ challenge is the promotion of systems and methodologies through the organization of a challenge on the tasks of large-scale biomedical semantic indexing and question answering. In total, 30 teams with more than 100 systems participated in the challenge this year. As in previous years, the best systems were able to outperform the strong baselines. This suggests that state-of-the-art systems are continuously improving, pushing the frontier of research.
* 17 pages, 2 figures
Semantic integration of disease-specific knowledge
Dec 18, 2019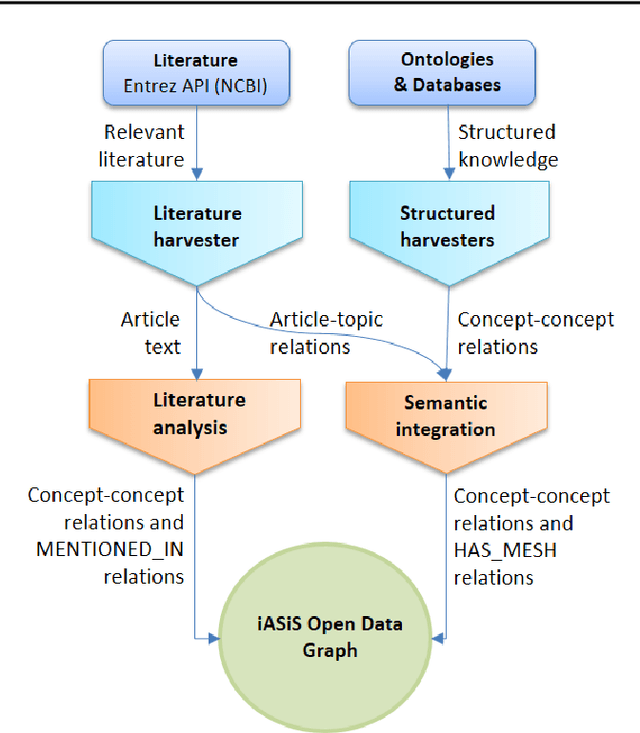
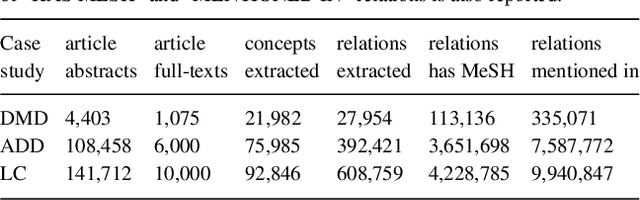

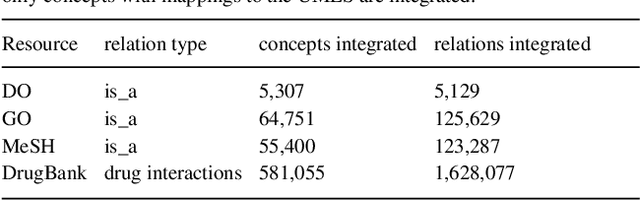
Abstract:Biomedical researchers working on a specific disease need up-to-date and unified access to knowledge relevant to the disease of their interest. Knowledge is continuously accumulated in scientific literature and other resources such as biomedical ontologies. Identifying the specific information needed is a challenging task and computational tools can be valuable. In this study, we propose a pipeline to automatically retrieve and integrate relevant knowledge based on a semantic graph representation, the iASiS Open Data Graph. Results: The disease-specific semantic graph can provide easy access to resources relevant to specific concepts and individual aspects of these concepts, in the form of concept relations and attributes. The proposed approach is applied to three different case studies: Two prevalent diseases, Lung Cancer and Dementia, for which a lot of knowledge is available, and one rare disease, Duchenne Muscular Dystrophy, for which knowledge is less abundant and difficult to locate. Results from exemplary queries are presented, investigating the potential of this approach in integrating and accessing knowledge as an automatically generated semantic graph.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge