Semantic integration of disease-specific knowledge
Paper and Code
Dec 18, 2019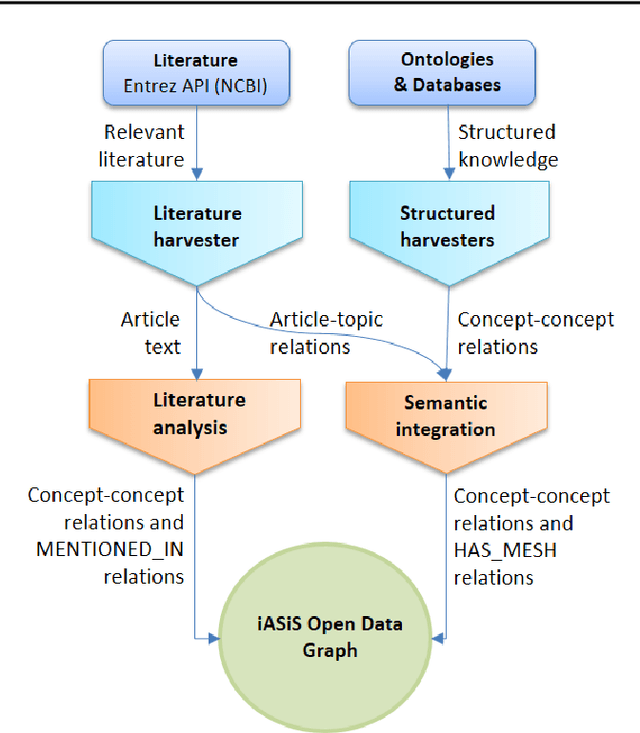
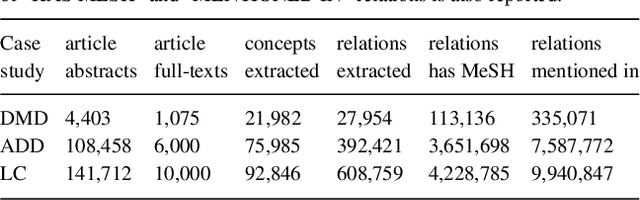

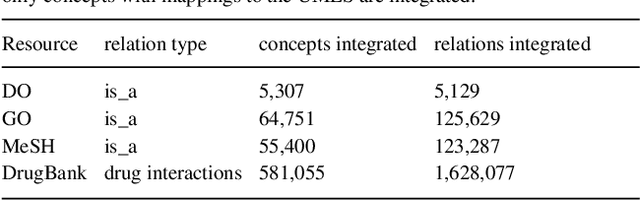
Biomedical researchers working on a specific disease need up-to-date and unified access to knowledge relevant to the disease of their interest. Knowledge is continuously accumulated in scientific literature and other resources such as biomedical ontologies. Identifying the specific information needed is a challenging task and computational tools can be valuable. In this study, we propose a pipeline to automatically retrieve and integrate relevant knowledge based on a semantic graph representation, the iASiS Open Data Graph. Results: The disease-specific semantic graph can provide easy access to resources relevant to specific concepts and individual aspects of these concepts, in the form of concept relations and attributes. The proposed approach is applied to three different case studies: Two prevalent diseases, Lung Cancer and Dementia, for which a lot of knowledge is available, and one rare disease, Duchenne Muscular Dystrophy, for which knowledge is less abundant and difficult to locate. Results from exemplary queries are presented, investigating the potential of this approach in integrating and accessing knowledge as an automatically generated semantic graph.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge