Anastasia Krithara
iASiS: Towards Heterogeneous Big Data Analysis for Personalized Medicine
Jul 09, 2024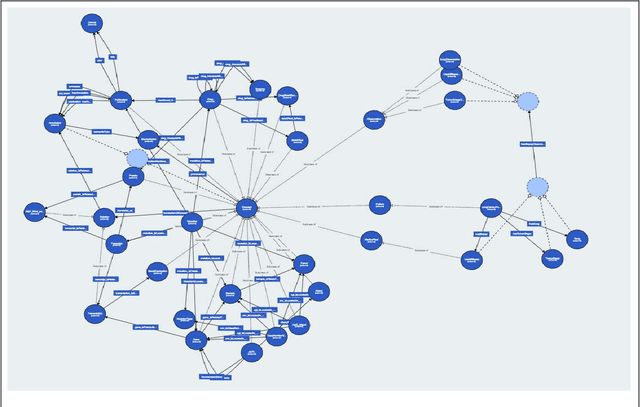
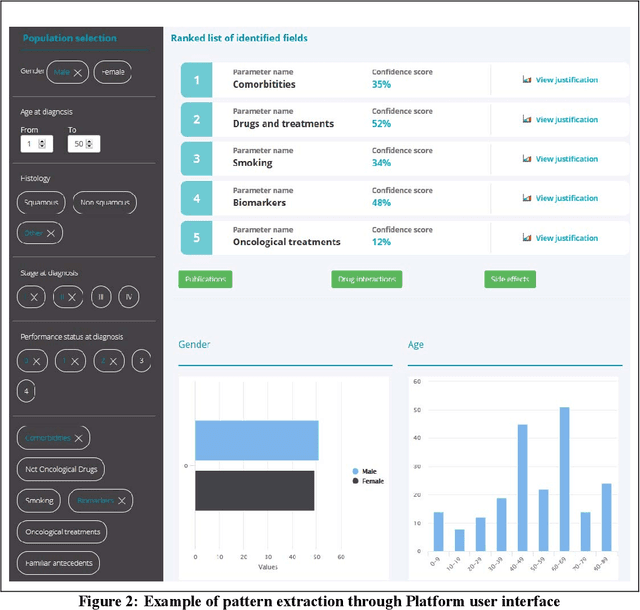
Abstract:The vision of IASIS project is to turn the wave of big biomedical data heading our way into actionable knowledge for decision makers. This is achieved by integrating data from disparate sources, including genomics, electronic health records and bibliography, and applying advanced analytics methods to discover useful patterns. The goal is to turn large amounts of available data into actionable information to authorities for planning public health activities and policies. The integration and analysis of these heterogeneous sources of information will enable the best decisions to be made, allowing for diagnosis and treatment to be personalised to each individual. The project offers a common representation schema for the heterogeneous data sources. The iASiS infrastructure is able to convert clinical notes into usable data, combine them with genomic data, related bibliography, image data and more, and create a global knowledge base. This facilitates the use of intelligent methods in order to discover useful patterns across different resources. Using semantic integration of data gives the opportunity to generate information that is rich, auditable and reliable. This information can be used to provide better care, reduce errors and create more confidence in sharing data, thus providing more insights and opportunities. Data resources for two different disease categories are explored within the iASiS use cases, dementia and lung cancer.
* 6 pages, 2 figures, accepted at 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS)
Overview of BioASQ 2023: The eleventh BioASQ challenge on Large-Scale Biomedical Semantic Indexing and Question Answering
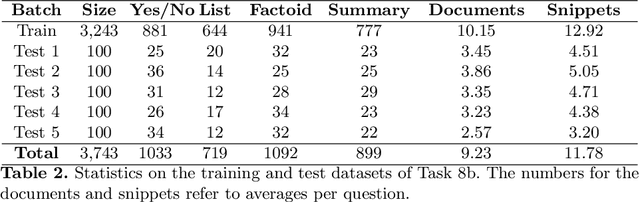
Jul 11, 2023Abstract:This is an overview of the eleventh edition of the BioASQ challenge in the context of the Conference and Labs of the Evaluation Forum (CLEF) 2023. BioASQ is a series of international challenges promoting advances in large-scale biomedical semantic indexing and question answering. This year, BioASQ consisted of new editions of the two established tasks b and Synergy, and a new task (MedProcNER) on semantic annotation of clinical content in Spanish with medical procedures, which have a critical role in medical practice. In this edition of BioASQ, 28 competing teams submitted the results of more than 150 distinct systems in total for the three different shared tasks of the challenge. Similarly to previous editions, most of the participating systems achieved competitive performance, suggesting the continuous advancement of the state-of-the-art in the field.
Large-scale fine-grained semantic indexing of biomedical literature based on weakly-supervised deep learning
Jan 23, 2023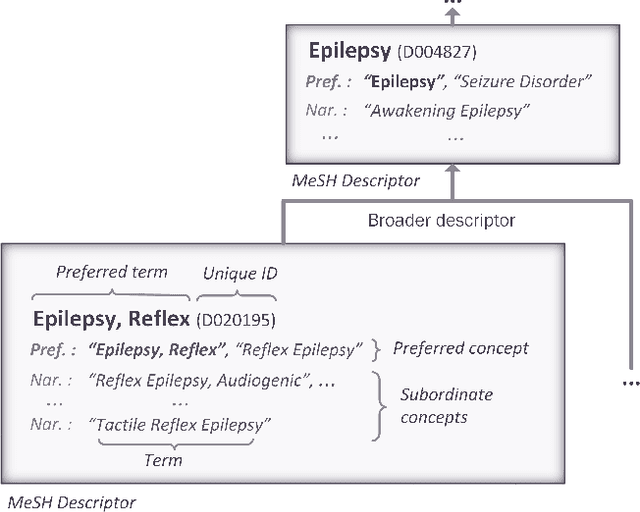
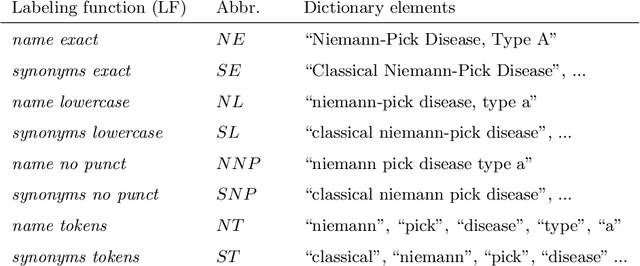
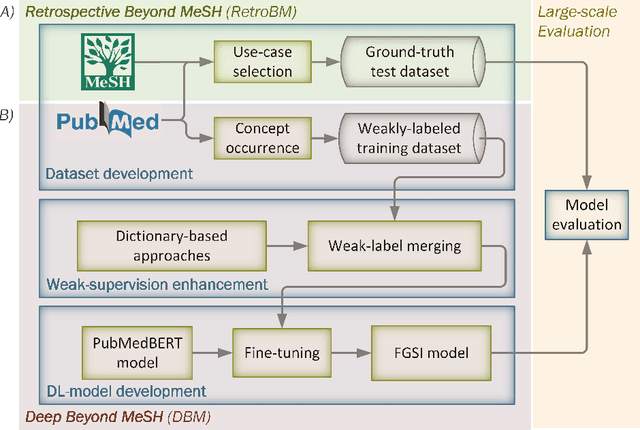
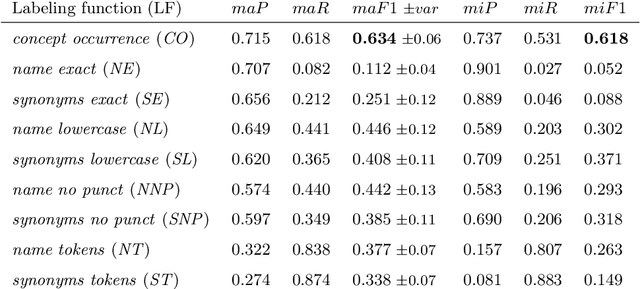
Abstract:Semantic indexing of biomedical literature is usually done at the level of MeSH descriptors, representing topics of interest for the biomedical community. Several related but distinct biomedical concepts are often grouped together in a single coarse-grained descriptor and are treated as a single topic for semantic indexing. This study proposes a new method for the automated refinement of subject annotations at the level of concepts, investigating deep learning approaches. Lacking labelled data for this task, our method relies on weak supervision based on concept occurrence in the abstract of an article. The proposed approach is evaluated on an extended large-scale retrospective scenario, taking advantage of concepts that eventually become MeSH descriptors, for which annotations become available in MEDLINE/PubMed. The results suggest that concept occurrence is a strong heuristic for automated subject annotation refinement and can be further enhanced when combined with dictionary-based heuristics. In addition, such heuristics can be useful as weak supervision for developing deep learning models that can achieve further improvement in some cases.
Overview of BioASQ 2022: The tenth BioASQ challenge on Large-Scale Biomedical Semantic Indexing and Question Answering
Oct 13, 2022Abstract:This paper presents an overview of the tenth edition of the BioASQ challenge in the context of the Conference and Labs of the Evaluation Forum (CLEF) 2022. BioASQ is an ongoing series of challenges that promotes advances in the domain of large-scale biomedical semantic indexing and question answering. In this edition, the challenge was composed of the three established tasks a, b, and Synergy, and a new task named DisTEMIST for automatic semantic annotation and grounding of diseases from clinical content in Spanish, a key concept for semantic indexing and search engines of literature and clinical records. This year, BioASQ received more than 170 distinct systems from 38 teams in total for the four different tasks of the challenge. As in previous years, the majority of the competing systems outperformed the strong baselines, indicating the continuous advancement of the state-of-the-art in this domain.
* 25 pages, 14 tables, 4 figures. arXiv admin note: substantial text overlap with arXiv:2106.14885
Overview of BioASQ 2020: The eighth BioASQ challenge on Large-Scale Biomedical Semantic Indexing and Question Answering
Jun 28, 2021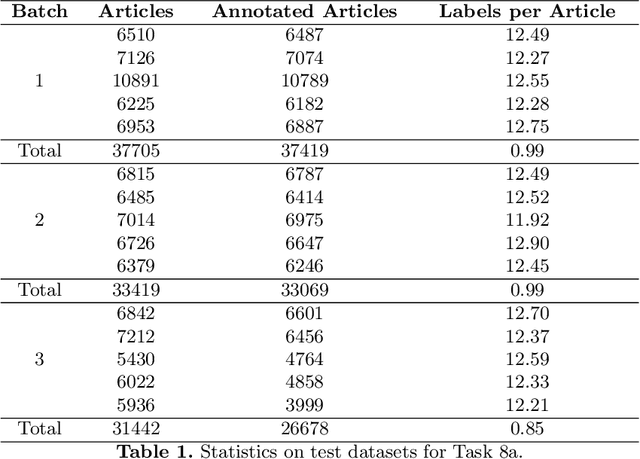
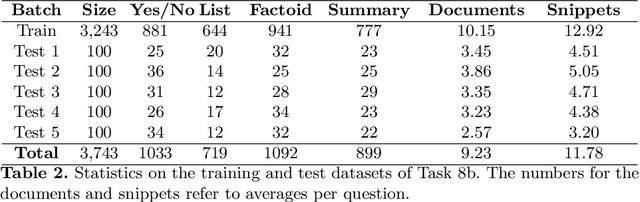
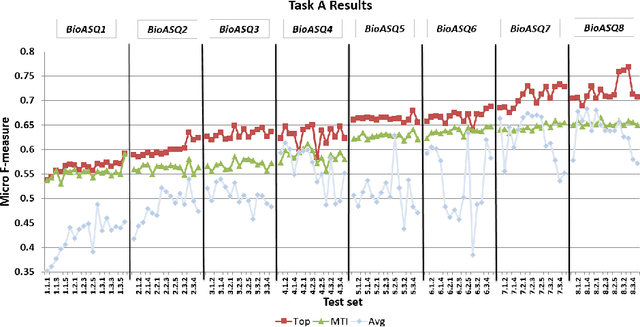
Abstract:In this paper, we present an overview of the eighth edition of the BioASQ challenge, which ran as a lab in the Conference and Labs of the Evaluation Forum (CLEF) 2020. BioASQ is a series of challenges aiming at the promotion of systems and methodologies for large-scale biomedical semantic indexing and question answering. To this end, shared tasks are organized yearly since 2012, where different teams develop systems that compete on the same demanding benchmark datasets that represent the real information needs of experts in the biomedical domain. This year, the challenge has been extended with the introduction of a new task on medical semantic indexing in Spanish. In total, 34 teams with more than 100 systems participated in the three tasks of the challenge. As in previous years, the results of the evaluation reveal that the top-performing systems managed to outperform the strong baselines, which suggests that state-of-the-art systems keep pushing the frontier of research through continuous improvements.
* 21 pages, 10 tables, 3 figures
Overview of BioASQ 2021: The ninth BioASQ challenge on Large-Scale Biomedical Semantic Indexing and Question Answering
Jun 28, 2021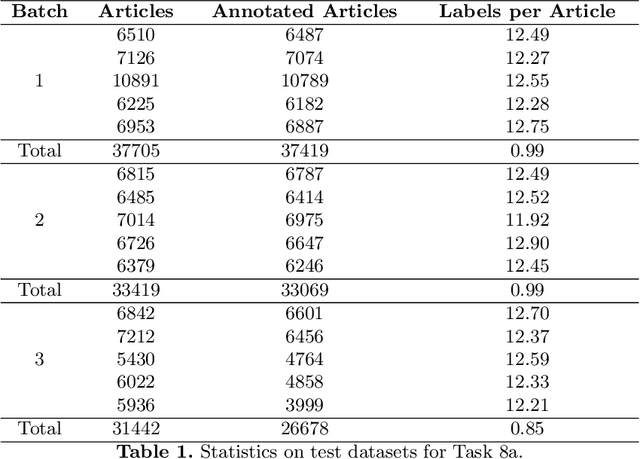
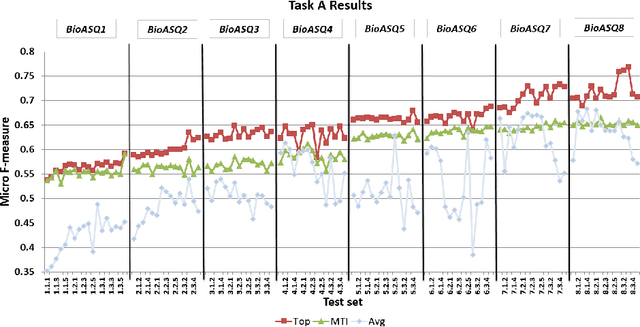
Abstract:Advancing the state-of-the-art in large-scale biomedical semantic indexing and question answering is the main focus of the BioASQ challenge. BioASQ organizes respective tasks where different teams develop systems that are evaluated on the same benchmark datasets that represent the real information needs of experts in the biomedical domain. This paper presents an overview of the ninth edition of the BioASQ challenge in the context of the Conference and Labs of the Evaluation Forum (CLEF) 2021. In this year, a new question answering task, named Synergy, is introduced to support researchers studying the COVID-19 disease and measure the ability of the participating teams to discern information while the problem is still developing. In total, 42 teams with more than 170 systems were registered to participate in the four tasks of the challenge. The evaluation results, similarly to previous years, show a performance gain against the baselines which indicates the continuous improvement of the state-of-the-art in this field.
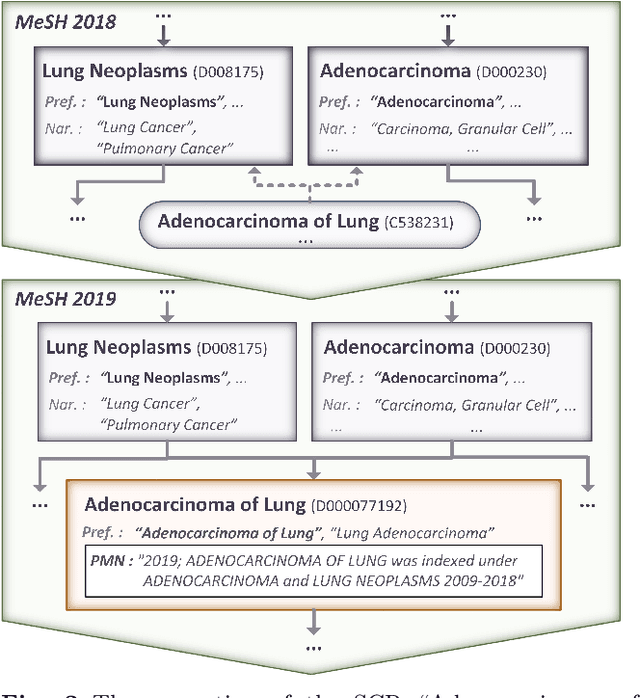
Harvesting the Public MeSH Note field
Jun 01, 2021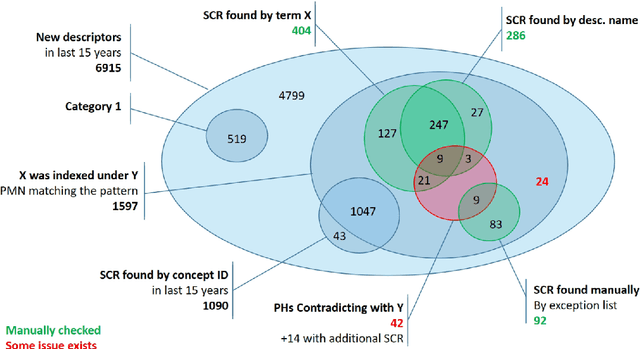

Abstract:In this document, we report an analysis of the Public MeSH Note field of the new descriptors introduced in the MeSH thesaurus between 2006 and 2020. The aim of this analysis was to extract information about the previous status of these new descriptors as Supplementary Concept Records. The Public MeSH Note field contains information in semi-structured text, meant to be read by humans. Therefore, we adopted a semi-automated approach, based on regular expressions, to extract information from it. In the large majority of cases, we managed to minimize the required manual effort for extracting the previous state of a new descriptor as a Supplementary Concept Record. The source code for this analysis is openly available on GitHub.
What is all this new MeSH about? Exploring the semantic provenance of new descriptors in the MeSH thesaurus
Jan 20, 2021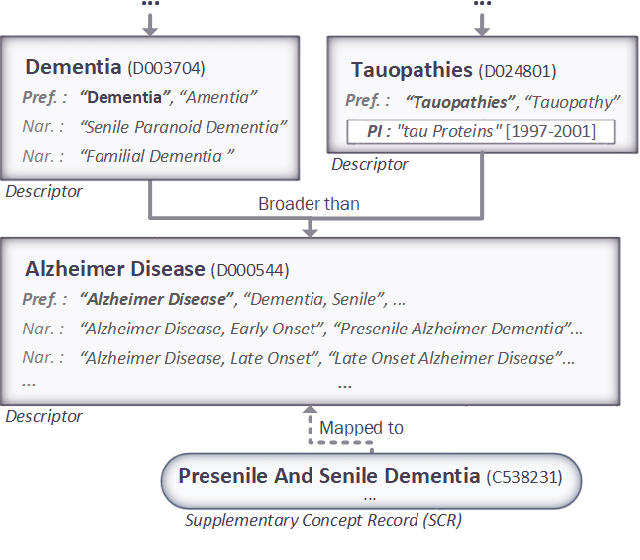
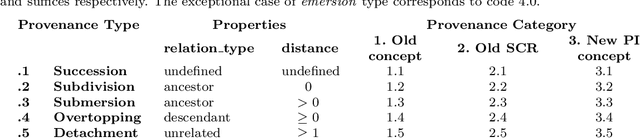
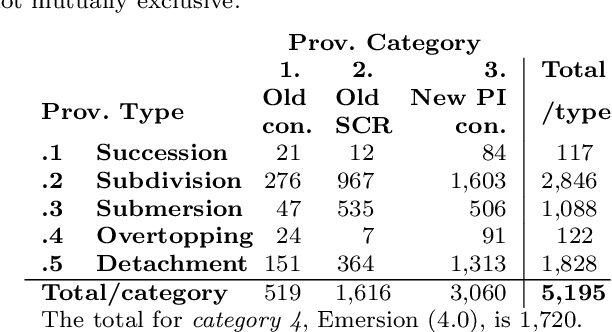
Abstract:The Medical Subject Headings (MeSH) thesaurus is a controlled vocabulary widely used in biomedical knowledge systems, particularly for semantic indexing of scientific literature. As the MeSH hierarchy evolves through annual version updates, some new descriptors are introduced that were not previously available. This paper explores the conceptual provenance of these new descriptors. In particular, we investigate whether such new descriptors have been previously covered by older descriptors and what is their current relation to them. To this end, we propose a framework to categorize new descriptors based on their current relation to older descriptors. Based on the proposed classification scheme, we quantify, analyse and present the different types of new descriptors introduced in MeSH during the last fifteen years. The results show that only about 25% of new MeSH descriptors correspond to new emerging concepts, whereas the rest were previously covered by one or more existing descriptors, either implicitly or explicitly. Most of them were covered by a single existing descriptor and they usually end up as descendants of it in the current hierarchy, gradually leading towards a more fine-grained MeSH vocabulary. These insights about the dynamics of the thesaurus are useful for the retrospective study of scientific articles annotated with MeSH, but could also be used to inform the policy of updating the thesaurus in the future.
Results of the seventh edition of the BioASQ Challenge
Jun 16, 2020
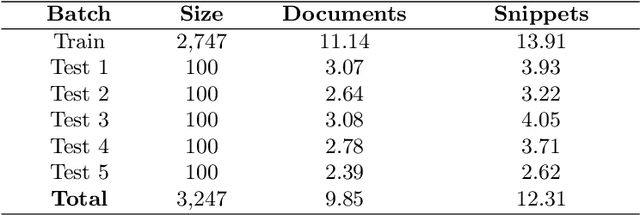
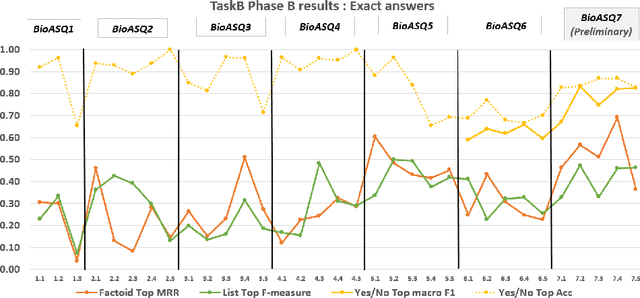
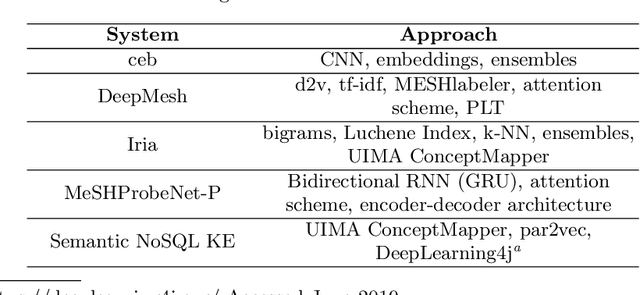
Abstract:The results of the seventh edition of the BioASQ challenge are presented in this paper. The aim of the BioASQ challenge is the promotion of systems and methodologies through the organization of a challenge on the tasks of large-scale biomedical semantic indexing and question answering. In total, 30 teams with more than 100 systems participated in the challenge this year. As in previous years, the best systems were able to outperform the strong baselines. This suggests that state-of-the-art systems are continuously improving, pushing the frontier of research.
* 17 pages, 2 figures
Semantic integration of disease-specific knowledge
Dec 18, 2019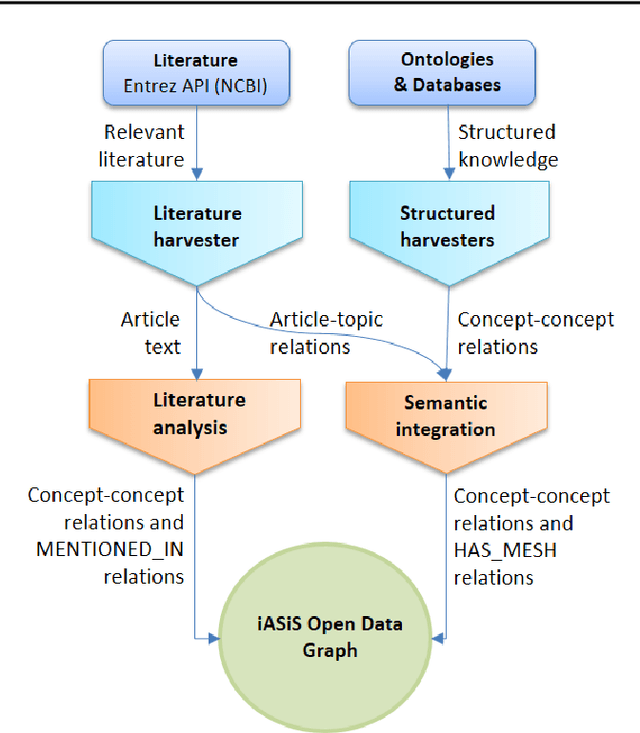
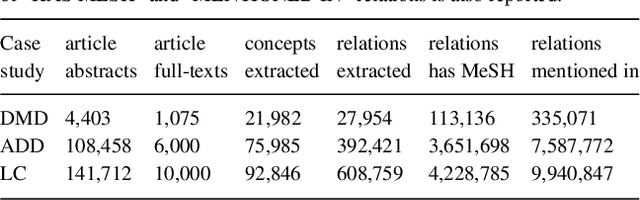

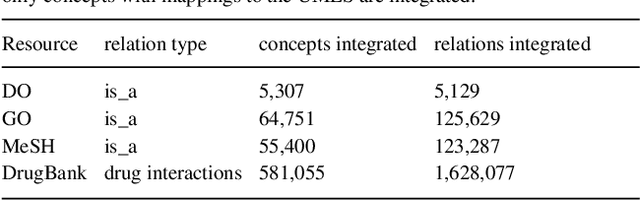
Abstract:Biomedical researchers working on a specific disease need up-to-date and unified access to knowledge relevant to the disease of their interest. Knowledge is continuously accumulated in scientific literature and other resources such as biomedical ontologies. Identifying the specific information needed is a challenging task and computational tools can be valuable. In this study, we propose a pipeline to automatically retrieve and integrate relevant knowledge based on a semantic graph representation, the iASiS Open Data Graph. Results: The disease-specific semantic graph can provide easy access to resources relevant to specific concepts and individual aspects of these concepts, in the form of concept relations and attributes. The proposed approach is applied to three different case studies: Two prevalent diseases, Lung Cancer and Dementia, for which a lot of knowledge is available, and one rare disease, Duchenne Muscular Dystrophy, for which knowledge is less abundant and difficult to locate. Results from exemplary queries are presented, investigating the potential of this approach in integrating and accessing knowledge as an automatically generated semantic graph.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge