Maria-Esther Vidal
Semantic Web: Past, Present, and Future
Dec 22, 2024Abstract:Ever since the vision was formulated, the Semantic Web has inspired many generations of innovations. Semantic technologies have been used to share vast amounts of information on the Web, enhance them with semantics to give them meaning, and enable inference and reasoning on them. Throughout the years, semantic technologies, and in particular knowledge graphs, have been used in search engines, data integration, enterprise settings, and machine learning. In this paper, we recap the classical concepts and foundations of the Semantic Web as well as modern and recent concepts and applications, building upon these foundations. The classical topics we cover include knowledge representation, creating and validating knowledge on the Web, reasoning and linking, and distributed querying. We enhance this classical view of the so-called ``Semantic Web Layer Cake'' with an update of recent concepts that include provenance, security and trust, as well as a discussion of practical impacts from industry-led contributions. We conclude with an outlook on the future directions of the Semantic Web.
* Extended Version 2024-12-13 of TGDK 2(1): 3:1-3:37 (2024) If you like to contribute, please contact the first author and visit: https://github.com/ascherp/semantic-web-primer Please cite this paper as, see https://dblp.org/rec/journals/tgdk/ScherpG0HV24.html?view=bibtex
Articulation Work and Tinkering for Fairness in Machine Learning
Jul 23, 2024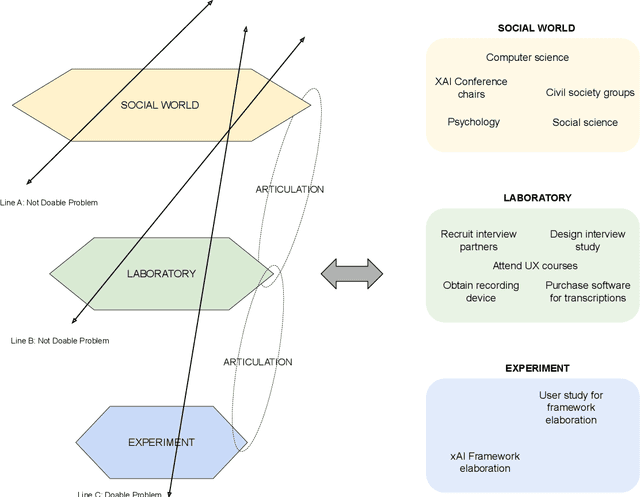
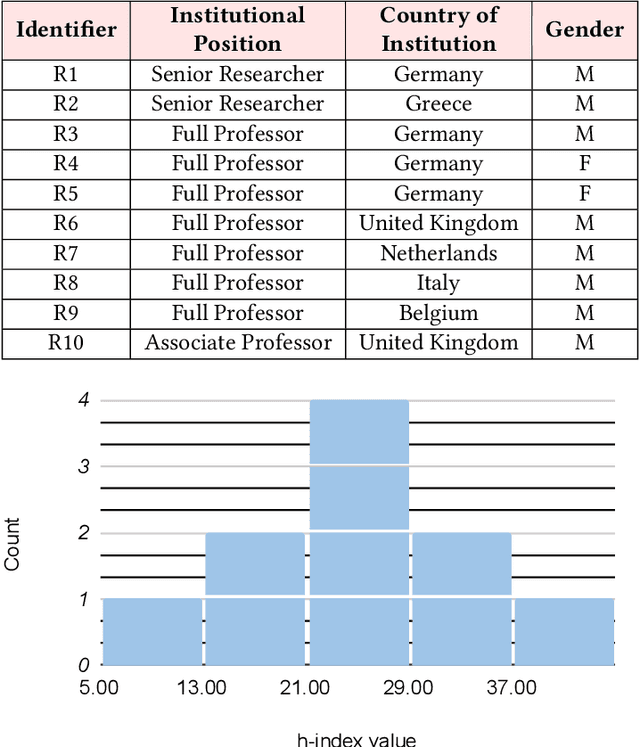
Abstract:The field of fair AI aims to counter biased algorithms through computational modelling. However, it faces increasing criticism for perpetuating the use of overly technical and reductionist methods. As a result, novel approaches appear in the field to address more socially-oriented and interdisciplinary (SOI) perspectives on fair AI. In this paper, we take this dynamic as the starting point to study the tension between computer science (CS) and SOI research. By drawing on STS and CSCW theory, we position fair AI research as a matter of 'organizational alignment': what makes research 'doable' is the successful alignment of three levels of work organization (the social world, the laboratory and the experiment). Based on qualitative interviews with CS researchers, we analyze the tasks, resources, and actors required for doable research in the case of fair AI. We find that CS researchers engage with SOI to some extent, but organizational conditions, articulation work, and ambiguities of the social world constrain the doability of SOI research. Based on our findings, we identify and discuss problems for aligning CS and SOI as fair AI continues to evolve.
iASiS: Towards Heterogeneous Big Data Analysis for Personalized Medicine
Jul 09, 2024
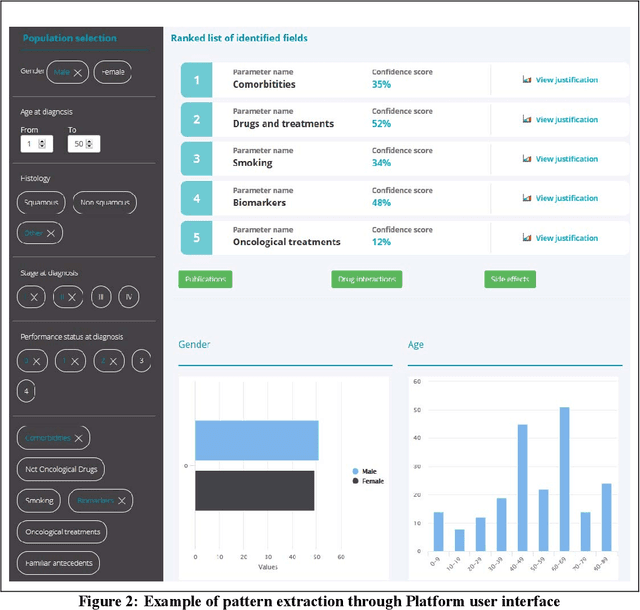
Abstract:The vision of IASIS project is to turn the wave of big biomedical data heading our way into actionable knowledge for decision makers. This is achieved by integrating data from disparate sources, including genomics, electronic health records and bibliography, and applying advanced analytics methods to discover useful patterns. The goal is to turn large amounts of available data into actionable information to authorities for planning public health activities and policies. The integration and analysis of these heterogeneous sources of information will enable the best decisions to be made, allowing for diagnosis and treatment to be personalised to each individual. The project offers a common representation schema for the heterogeneous data sources. The iASiS infrastructure is able to convert clinical notes into usable data, combine them with genomic data, related bibliography, image data and more, and create a global knowledge base. This facilitates the use of intelligent methods in order to discover useful patterns across different resources. Using semantic integration of data gives the opportunity to generate information that is rich, auditable and reliable. This information can be used to provide better care, reduce errors and create more confidence in sharing data, thus providing more insights and opportunities. Data resources for two different disease categories are explored within the iASiS use cases, dementia and lung cancer.
* 6 pages, 2 figures, accepted at 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS)
Leveraging Ontologies to Document Bias in Data
Jun 29, 2024Abstract:Machine Learning (ML) systems are capable of reproducing and often amplifying undesired biases. This puts emphasis on the importance of operating under practices that enable the study and understanding of the intrinsic characteristics of ML pipelines, prompting the emergence of documentation frameworks with the idea that ``any remedy for bias starts with awareness of its existence''. However, a resource that can formally describe these pipelines in terms of biases detected is still amiss. To fill this gap, we present the Doc-BiasO ontology, a resource that aims to create an integrated vocabulary of biases defined in the \textit{fair-ML} literature and their measures, as well as to incorporate relevant terminology and the relationships between them. Overseeing ontology engineering best practices, we re-use existing vocabulary on machine learning and AI, to foster knowledge sharing and interoperability between the actors concerned with its research, development, regulation, among others. Overall, our main objective is to contribute towards clarifying existing terminology on bias research as it rapidly expands to all areas of AI and to improve the interpretation of bias in data and downstream impact.
Traditional Machine Learning Models and Bidirectional Encoder Representations From Transformer (BERT)-Based Automatic Classification of Tweets About Eating Disorders: Algorithm Development and Validation Study
Feb 08, 2024



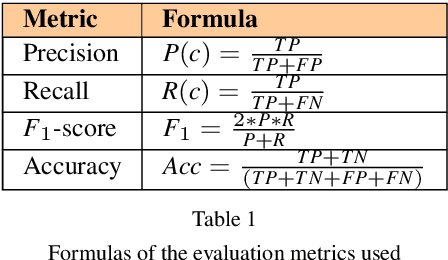
Abstract:Background: Eating disorders are increasingly prevalent, and social networks offer valuable information. Objective: Our goal was to identify efficient machine learning models for categorizing tweets related to eating disorders. Methods: Over three months, we collected tweets about eating disorders. A 2,000-tweet subset was labeled for: (1) being written by individuals with eating disorders, (2) promoting eating disorders, (3) informativeness, and (4) scientific content. Both traditional machine learning and deep learning models were employed for classification, assessing accuracy, F1 score, and computational time. Results: From 1,058,957 collected tweets, transformer-based bidirectional encoder representations achieved the highest F1 scores (71.1%-86.4%) across all four categories. Conclusions: Transformer-based models outperform traditional techniques in classifying eating disorder-related tweets, though they require more computational resources.
Empowering machine learning models with contextual knowledge for enhancing the detection of eating disorders in social media posts
Feb 08, 2024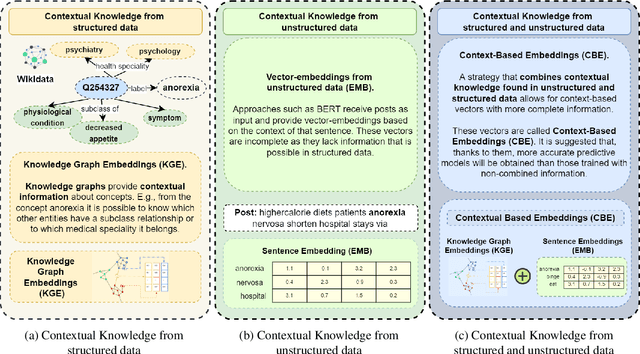
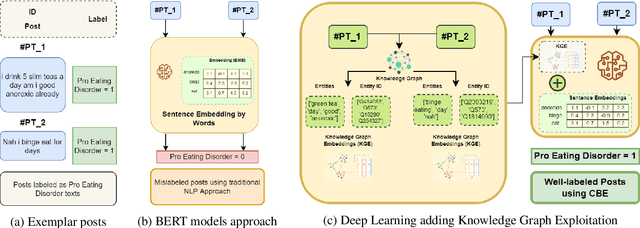
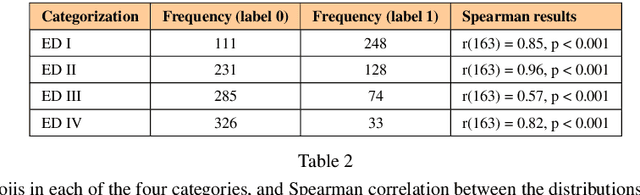
Abstract:Social networks are vital for information sharing, especially in the health sector for discussing diseases and treatments. These platforms, however, often feature posts as brief texts, posing challenges for Artificial Intelligence (AI) in understanding context. We introduce a novel hybrid approach combining community-maintained knowledge graphs (like Wikidata) with deep learning to enhance the categorization of social media posts. This method uses advanced entity recognizers and linkers (like Falcon 2.0) to connect short post entities to knowledge graphs. Knowledge graph embeddings (KGEs) and contextualized word embeddings (like BERT) are then employed to create rich, context-based representations of these posts. Our focus is on the health domain, particularly in identifying posts related to eating disorders (e.g., anorexia, bulimia) to aid healthcare providers in early diagnosis. We tested our approach on a dataset of 2,000 tweets about eating disorders, finding that merging word embeddings with knowledge graph information enhances the predictive models' reliability. This methodology aims to assist health experts in spotting patterns indicative of mental disorders, thereby improving early detection and accurate diagnosis for personalized medicine.
Trust, Accountability, and Autonomy in Knowledge Graph-based AI for Self-determination
Oct 31, 2023Abstract:Knowledge Graphs (KGs) have emerged as fundamental platforms for powering intelligent decision-making and a wide range of Artificial Intelligence (AI) services across major corporations such as Google, Walmart, and AirBnb. KGs complement Machine Learning (ML) algorithms by providing data context and semantics, thereby enabling further inference and question-answering capabilities. The integration of KGs with neuronal learning (e.g., Large Language Models (LLMs)) is currently a topic of active research, commonly named neuro-symbolic AI. Despite the numerous benefits that can be accomplished with KG-based AI, its growing ubiquity within online services may result in the loss of self-determination for citizens as a fundamental societal issue. The more we rely on these technologies, which are often centralised, the less citizens will be able to determine their own destinies. To counter this threat, AI regulation, such as the European Union (EU) AI Act, is being proposed in certain regions. The regulation sets what technologists need to do, leading to questions concerning: How can the output of AI systems be trusted? What is needed to ensure that the data fuelling and the inner workings of these artefacts are transparent? How can AI be made accountable for its decision-making? This paper conceptualises the foundational topics and research pillars to support KG-based AI for self-determination. Drawing upon this conceptual framework, challenges and opportunities for citizen self-determination are illustrated and analysed in a real-world scenario. As a result, we propose a research agenda aimed at accomplishing the recommended objectives.
Scaling Up Knowledge Graph Creation to Large and Heterogeneous Data Sources
Jan 24, 2022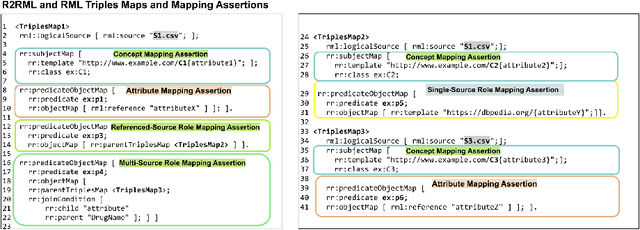

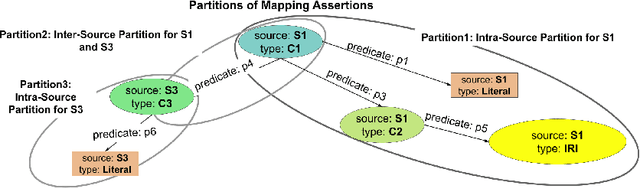
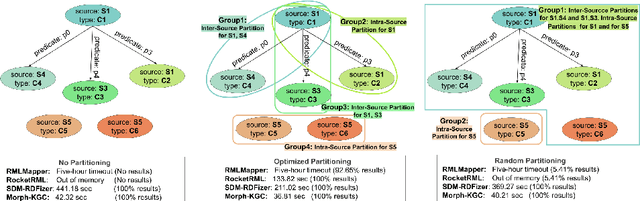
Abstract:RDF knowledge graphs (KG) are powerful data structures to represent factual statements created from heterogeneous data sources. KG creation is laborious, and demands data management techniques to be executed efficiently. This paper tackles the problem of the automatic generation of KG creation processes declaratively specified; it proposes techniques for planning and transforming heterogeneous data into RDF triples following mapping assertions specified in the RDF Mapping Language (RML). Given a set of mapping assertions, the planner provides an optimized execution plan by partitioning and scheduling the execution of the assertions. First, the planner assesses an optimized number of partitions considering the number of data sources, type of mapping assertions, and the associations between different assertions. After providing a list of partitions and assertions that belong to each partition, the planner determines their execution order. A greedy algorithm is implemented to generate the partitions' bushy tree execution plan. Bushy tree plans are translated into operating system commands that guide the execution of the partitions of the mapping assertions in the order indicated by the bushy tree. The proposed optimization approach is evaluated over state-of-the-art RML-compliant engines and existing benchmarks of data sources and RML triples maps. Our experimental results suggest that the performance of the studied engines can be considerably improved, particularly in a complex setting with numerous triples maps and data sources. As a result, engines that usually time in complex cases out can, if not entirely execute all the assertions, still produce a portion of the KG.
EABlock: A Declarative Entity Alignment Block for Knowledge Graph Creation Pipelines
Dec 15, 2021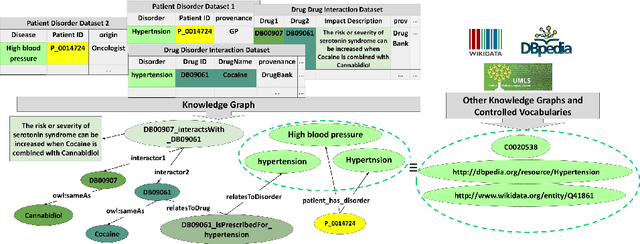
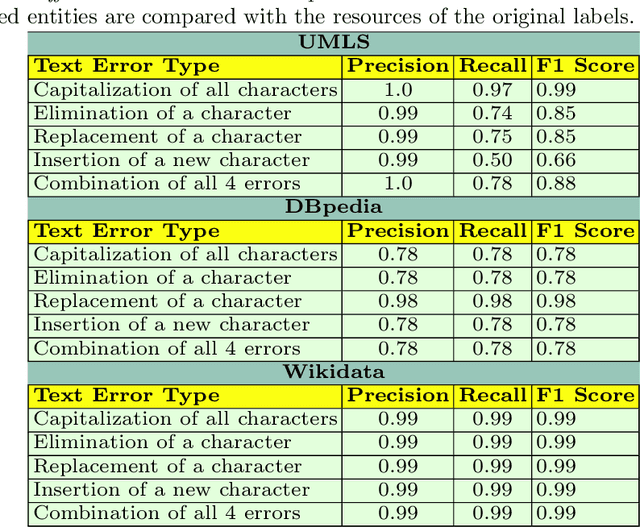
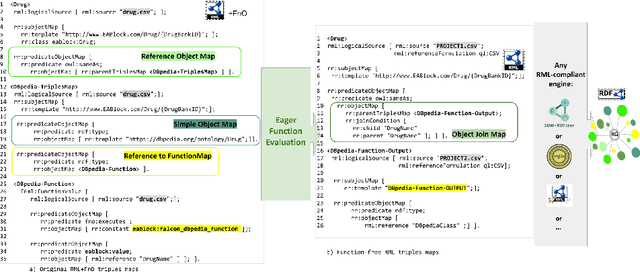
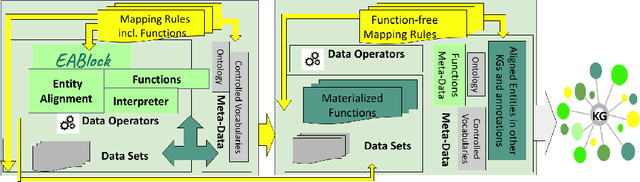
Abstract:Despite encoding enormous amount of rich and valuable data, existing data sources are mostly created independently, being a significant challenge to their integration. Mapping languages, e.g., RML and R2RML, facilitate declarative specification of the process of applying meta-data and integrating data into a knowledge graph. Mapping rules can also include knowledge extraction functions in addition to expressing correspondences among data sources and a unified schema. Combining mapping rules and functions represents a powerful formalism to specify pipelines for integrating data into a knowledge graph transparently. Surprisingly, these formalisms are not fully adapted, and many knowledge graphs are created by executing ad-hoc programs to pre-process and integrate data. In this paper, we present EABlock, an approach integrating Entity Alignment (EA) as part of RML mapping rules. EABlock includes a block of functions performing entity recognition from textual attributes and link the recognized entities to the corresponding resources in Wikidata, DBpedia, and domain specific thesaurus, e.g., UMLS. EABlock provides agnostic and efficient techniques to evaluate the functions and transfer the mappings to facilitate its application in any RML-compliant engine. We have empirically evaluated EABlock performance, and results indicate that EABlock speeds up knowledge graph creation pipelines that require entity recognition and linking in state-of-the-art RML-compliant engines. EABlock is also publicly available as a tool through a GitHub repository(https://github.com/SDM-TIB/EABlock) and a DOI(https://doi.org/10.5281/zenodo.5779773).
Unveiling Relations in the Industry 4.0 Standards Landscape based on Knowledge Graph Embeddings
Jun 03, 2020
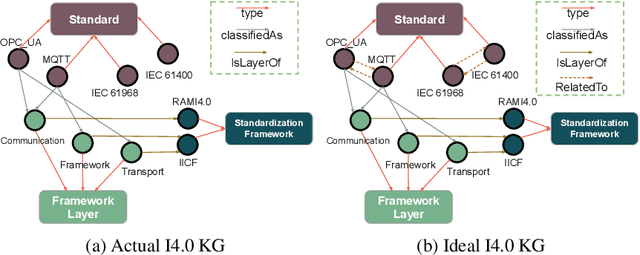
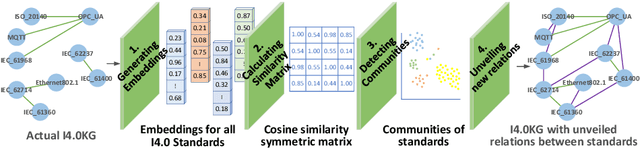
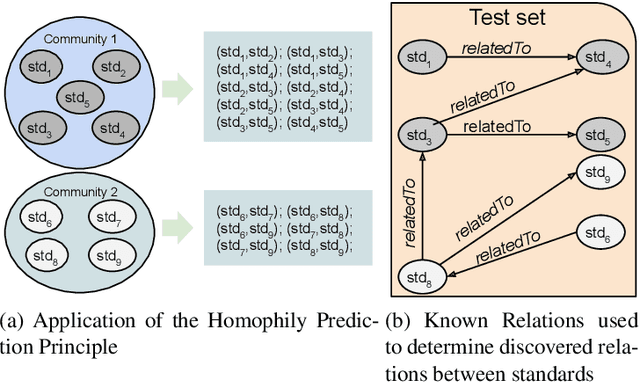
Abstract:Industry~4.0 (I4.0) standards and standardization frameworks have been proposed with the goal of \emph{empowering interoperability} in smart factories. These standards enable the description and interaction of the main components, systems, and processes inside of a smart factory. Due to the growing number of frameworks and standards, there is an increasing need for approaches that automatically analyze the landscape of I4.0 standards. Standardization frameworks classify standards according to their functions into layers and dimensions. However, similar standards can be classified differently across the frameworks, producing, thus, interoperability conflicts among them. Semantic-based approaches that rely on ontologies and knowledge graphs, have been proposed to represent standards, known relations among them, as well as their classification according to existing frameworks. Albeit informative, the structured modeling of the I4.0 landscape only provides the foundations for detecting interoperability issues. Thus, graph-based analytical methods able to exploit knowledge encoded by these approaches, are required to uncover alignments among standards. We study the relatedness among standards and frameworks based on community analysis to discover knowledge that helps to cope with interoperability conflicts between standards. We use knowledge graph embeddings to automatically create these communities exploiting the meaning of the existing relationships. In particular, we focus on the identification of similar standards, i.e., communities of standards, and analyze their properties to detect unknown relations. We empirically evaluate our approach on a knowledge graph of I4.0 standards using the Trans$^*$ family of embedding models for knowledge graph entities. Our results are promising and suggest that relations among standards can be detected accurately.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge