Ernestina Menasalvas
iASiS: Towards Heterogeneous Big Data Analysis for Personalized Medicine
Jul 09, 2024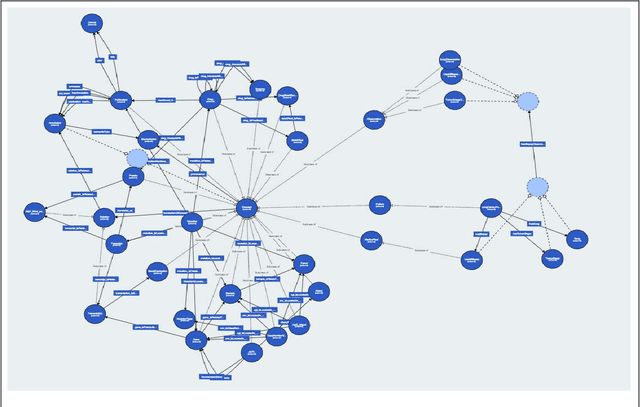
Abstract:The vision of IASIS project is to turn the wave of big biomedical data heading our way into actionable knowledge for decision makers. This is achieved by integrating data from disparate sources, including genomics, electronic health records and bibliography, and applying advanced analytics methods to discover useful patterns. The goal is to turn large amounts of available data into actionable information to authorities for planning public health activities and policies. The integration and analysis of these heterogeneous sources of information will enable the best decisions to be made, allowing for diagnosis and treatment to be personalised to each individual. The project offers a common representation schema for the heterogeneous data sources. The iASiS infrastructure is able to convert clinical notes into usable data, combine them with genomic data, related bibliography, image data and more, and create a global knowledge base. This facilitates the use of intelligent methods in order to discover useful patterns across different resources. Using semantic integration of data gives the opportunity to generate information that is rich, auditable and reliable. This information can be used to provide better care, reduce errors and create more confidence in sharing data, thus providing more insights and opportunities. Data resources for two different disease categories are explored within the iASiS use cases, dementia and lung cancer.
* 6 pages, 2 figures, accepted at 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS)
Machine Learning-Assisted Recurrence Prediction for Early-Stage Non-Small-Cell Lung Cancer Patients
Nov 17, 2022



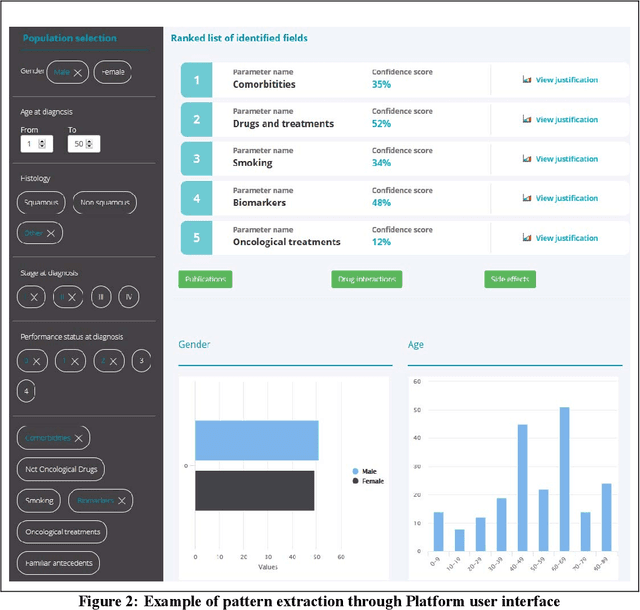
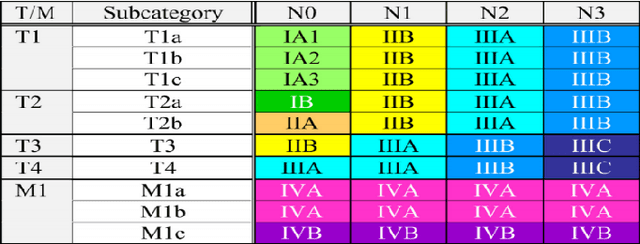
Abstract:Background: Stratifying cancer patients according to risk of relapse can personalize their care. In this work, we provide an answer to the following research question: How to utilize machine learning to estimate probability of relapse in early-stage non-small-cell lung cancer patients? Methods: For predicting relapse in 1,387 early-stage (I-II), non-small-cell lung cancer (NSCLC) patients from the Spanish Lung Cancer Group data (65.7 average age, 24.8% females, 75.2% males) we train tabular and graph machine learning models. We generate automatic explanations for the predictions of such models. For models trained on tabular data, we adopt SHAP local explanations to gauge how each patient feature contributes to the predicted outcome. We explain graph machine learning predictions with an example-based method that highlights influential past patients. Results: Machine learning models trained on tabular data exhibit a 76% accuracy for the Random Forest model at predicting relapse evaluated with a 10-fold cross-validation (model was trained 10 times with different independent sets of patients in test, train and validation sets, the reported metrics are averaged over these 10 test sets). Graph machine learning reaches 68% accuracy over a 200-patient, held-out test set, calibrated on a held-out set of 100 patients. Conclusions: Our results show that machine learning models trained on tabular and graph data can enable objective, personalised and reproducible prediction of relapse and therefore, disease outcome in patients with early-stage NSCLC. With further prospective and multisite validation, and additional radiological and molecular data, this prognostic model could potentially serve as a predictive decision support tool for deciding the use of adjuvant treatments in early-stage lung cancer. Keywords: Non-Small-Cell Lung Cancer, Tumor Recurrence Prediction, Machine Learning
Lung Cancer Concept Annotation from Spanish Clinical Narratives
Sep 18, 2018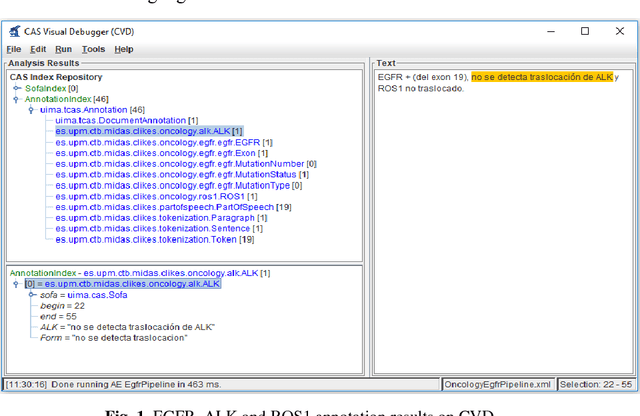
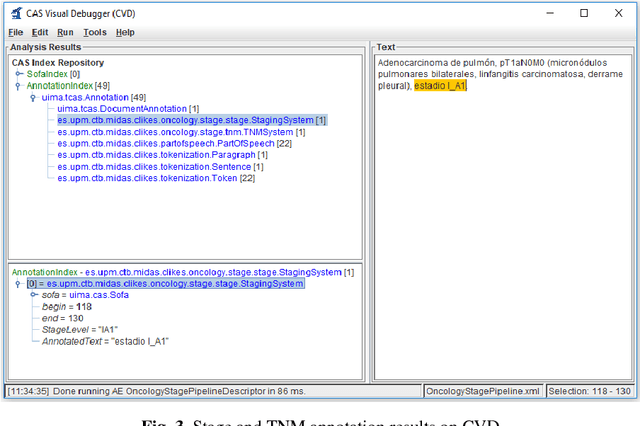
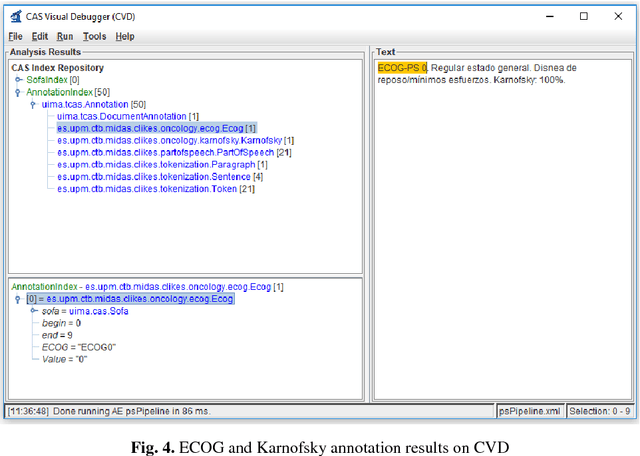
Abstract:Recent rapid increase in the generation of clinical data and rapid development of computational science make us able to extract new insights from massive datasets in healthcare industry. Oncological clinical notes are creating rich databases for documenting patients history and they potentially contain lots of patterns that could help in better management of the disease. However, these patterns are locked within free text (unstructured) portions of clinical documents and consequence in limiting health professionals to extract useful information from them and to finally perform Query and Answering (QA) process in an accurate way. The Information Extraction (IE) process requires Natural Language Processing (NLP) techniques to assign semantics to these patterns. Therefore, in this paper, we analyze the design of annotators for specific lung cancer concepts that can be integrated over Apache Unstructured Information Management Architecture (UIMA) framework. In addition, we explain the details of generation and storage of annotation outcomes.
* 10 pages, 6 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge