Alejandro Rodríguez-González
A Weak Supervision Approach for Monitoring Recreational Drug Use Effects in Social Media
Sep 18, 2025Abstract:Understanding the real-world effects of recreational drug use remains a critical challenge in public health and biomedical research, especially as traditional surveillance systems often underrepresent user experiences. In this study, we leverage social media (specifically Twitter) as a rich and unfiltered source of user-reported effects associated with three emerging psychoactive substances: ecstasy, GHB, and 2C-B. By combining a curated list of slang terms with biomedical concept extraction via MetaMap, we identified and weakly annotated over 92,000 tweets mentioning these substances. Each tweet was labeled with a polarity reflecting whether it reported a positive or negative effect, following an expert-guided heuristic process. We then performed descriptive and comparative analyses of the reported phenotypic outcomes across substances and trained multiple machine learning classifiers to predict polarity from tweet content, accounting for strong class imbalance using techniques such as cost-sensitive learning and synthetic oversampling. The top performance on the test set was obtained from eXtreme Gradient Boosting with cost-sensitive learning (F1 = 0.885, AUPRC = 0.934). Our findings reveal that Twitter enables the detection of substance-specific phenotypic effects, and that polarity classification models can support real-time pharmacovigilance and drug effect characterization with high accuracy.
A Semantic Social Network Analysis Tool for Sensitivity Analysis and What-If Scenario Testing in Alcohol Consumption Studies
Feb 14, 2024



Abstract:Social Network Analysis (SNA) is a set of techniques developed in the field of social and behavioral sciences research, in order to characterize and study the social relationships that are established among a set of individuals. When building a social network for performing an SNA analysis, an initial process of data gathering is achieved in order to extract the characteristics of the individuals and their relationships. This is usually done by completing a questionnaire containing different types of questions that will be later used to obtain the SNA measures needed to perform the study. There are, then, a great number of different possible network generating questions and also many possibilities for mapping the responses to the corresponding characteristics and relationships. Many variations may be introduced into these questions (the way they are posed, the weights given to each of the responses, etc.) that may have an effect on the resulting networks. All these different variations are difficult to achieve manually, because the process is time-consuming and error prone. The tool described in this paper uses semantic knowledge representation techniques in order to facilitate this kind of sensitivity studies. The base of the tool is a conceptual structure, called "ontology" that is able to represent the different concepts and their definitions. The tool is compared to other similar ones, and the advantages of the approach are highlighted, giving some particular examples from an ongoing SNA study about alcohol consumption habits in adolescents.
Social network analysis for personalized characterization and risk assessment of alcohol use disorders in adolescents using semantic technologies
Feb 14, 2024



Abstract:Alcohol Use Disorder (AUD) is a major concern for public health organizations worldwide, especially as regards the adolescent population. The consumption of alcohol in adolescents is known to be influenced by seeing friends and even parents drinking alcohol. Building on this fact, a number of studies into alcohol consumption among adolescents have made use of Social Network Analysis (SNA) techniques to study the different social networks (peers, friends, family, etc.) with whom the adolescent is involved. These kinds of studies need an initial phase of data gathering by means of questionnaires and a subsequent analysis phase using the SNA techniques. The process involves a number of manual data handling stages that are time consuming and error-prone. The use of knowledge engineering techniques (including the construction of a domain ontology) to represent the information, allows the automation of all the activities, from the initial data collection to the results of the SNA study. This paper shows how a knowledge model is constructed, and compares the results obtained using the traditional method with this, fully automated model, detailing the main advantages of the latter. In the case of the SNA analysis, the validity of the results obtained with the knowledge engineering approach are compared to those obtained manually using the UCINET, Cytoscape, Pajek and Gephi to test the accuracy of the knowledge model.
A new algorithm for Subgroup Set Discovery based on Information Gain
Jul 31, 2023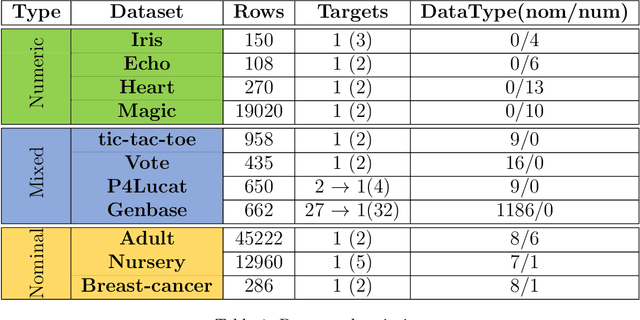

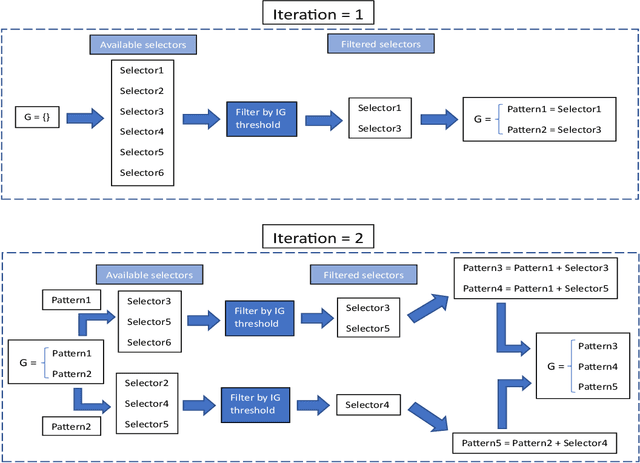
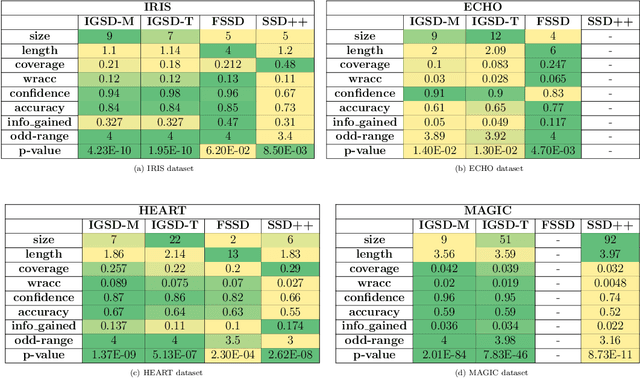
Abstract:Pattern discovery is a machine learning technique that aims to find sets of items, subsequences, or substructures that are present in a dataset with a higher frequency value than a manually set threshold. This process helps to identify recurring patterns or relationships within the data, allowing for valuable insights and knowledge extraction. In this work, we propose Information Gained Subgroup Discovery (IGSD), a new SD algorithm for pattern discovery that combines Information Gain (IG) and Odds Ratio (OR) as a multi-criteria for pattern selection. The algorithm tries to tackle some limitations of state-of-the-art SD algorithms like the need for fine-tuning of key parameters for each dataset, usage of a single pattern search criteria set by hand, usage of non-overlapping data structures for subgroup space exploration, and the impossibility to search for patterns by fixing some relevant dataset variables. Thus, we compare the performance of IGSD with two state-of-the-art SD algorithms: FSSD and SSD++. Eleven datasets are assessed using these algorithms. For the performance evaluation, we also propose to complement standard SD measures with IG, OR, and p-value. Obtained results show that FSSD and SSD++ algorithms provide less reliable patterns and reduced sets of patterns than IGSD algorithm for all datasets considered. Additionally, IGSD provides better OR values than FSSD and SSD++, stating a higher dependence between patterns and targets. Moreover, patterns obtained for one of the datasets used, have been validated by a group of domain experts. Thus, patterns provided by IGSD show better agreement with experts than patterns obtained by FSSD and SSD++ algorithms. These results demonstrate the suitability of the IGSD as a method for pattern discovery and suggest that the inclusion of non-standard SD metrics allows to better evaluate discovered patterns.
EBOCA: Evidences for BiOmedical Concepts Association Ontology
Aug 01, 2022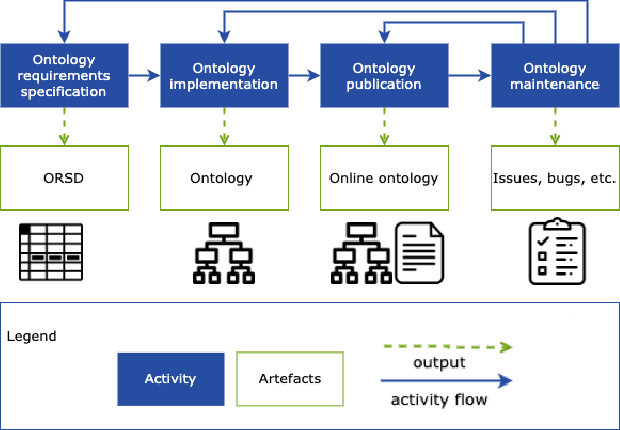
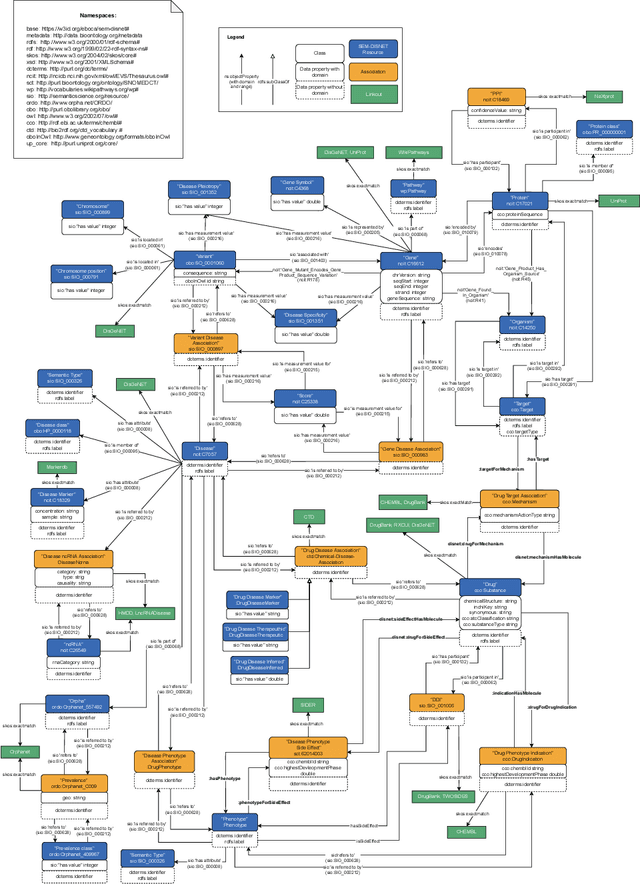
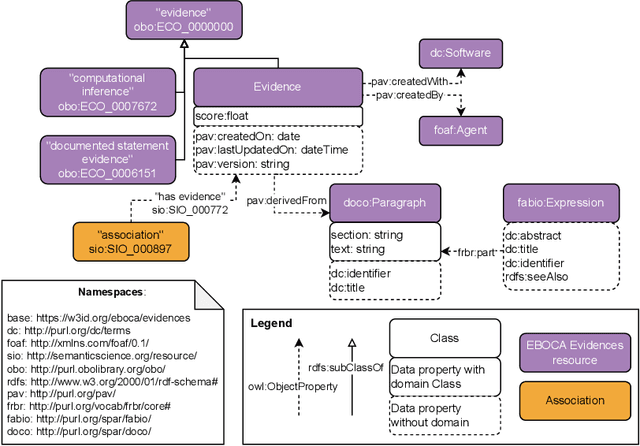
Abstract:There is a large number of online documents data sources available nowadays. The lack of structure and the differences between formats are the main difficulties to automatically extract information from them, which also has a negative impact on its use and reuse. In the biomedical domain, the DISNET platform emerged to provide researchers with a resource to obtain information in the scope of human disease networks by means of large-scale heterogeneous sources. Specifically in this domain, it is critical to offer not only the information extracted from different sources, but also the evidence that supports it. This paper proposes EBOCA, an ontology that describes (i) biomedical domain concepts and associations between them, and (ii) evidences supporting these associations; with the objective of providing an schema to improve the publication and description of evidences and biomedical associations in this domain. The ontology has been successfully evaluated to ensure there are no errors, modelling pitfalls and that it meets the previously defined functional requirements. Test data coming from a subset of DISNET and automatic association extractions from texts has been transformed according to the proposed ontology to create a Knowledge Graph that can be used in real scenarios, and which has also been used for the evaluation of the presented ontology.
Lung Cancer Concept Annotation from Spanish Clinical Narratives
Sep 18, 2018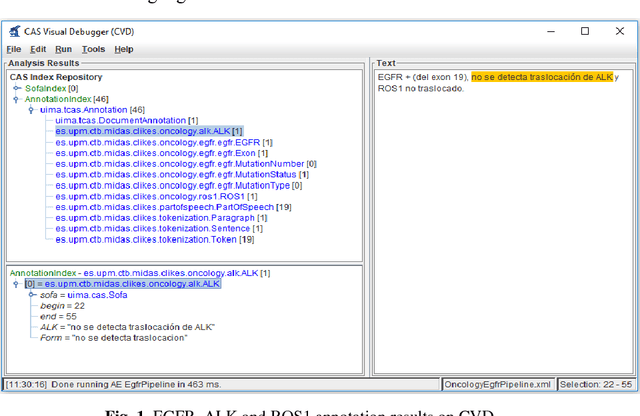
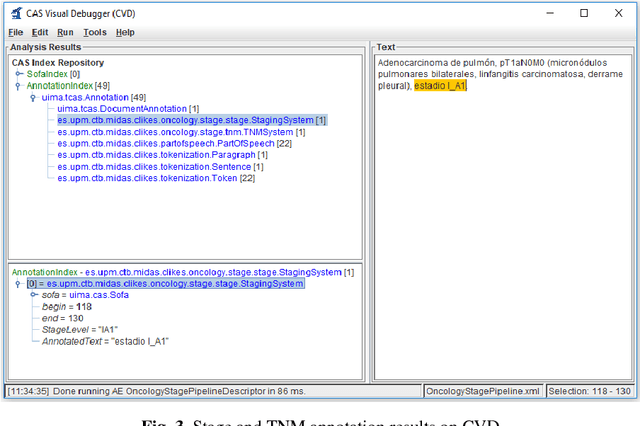
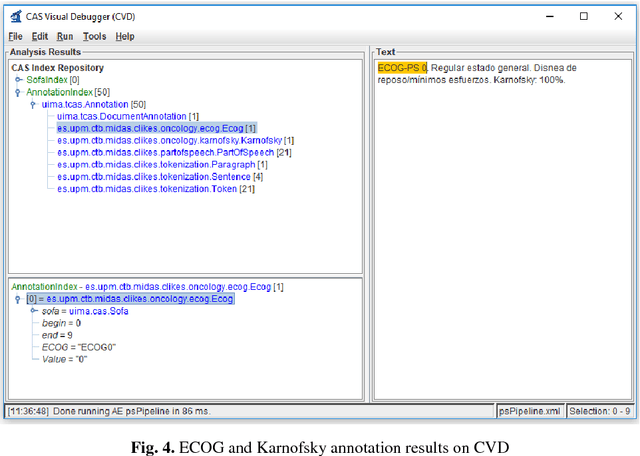
Abstract:Recent rapid increase in the generation of clinical data and rapid development of computational science make us able to extract new insights from massive datasets in healthcare industry. Oncological clinical notes are creating rich databases for documenting patients history and they potentially contain lots of patterns that could help in better management of the disease. However, these patterns are locked within free text (unstructured) portions of clinical documents and consequence in limiting health professionals to extract useful information from them and to finally perform Query and Answering (QA) process in an accurate way. The Information Extraction (IE) process requires Natural Language Processing (NLP) techniques to assign semantics to these patterns. Therefore, in this paper, we analyze the design of annotators for specific lung cancer concepts that can be integrated over Apache Unstructured Information Management Architecture (UIMA) framework. In addition, we explain the details of generation and storage of annotation outcomes.
* 10 pages, 6 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge