Carlos Badenes-Olmedo
EBOCA: Evidences for BiOmedical Concepts Association Ontology
Aug 01, 2022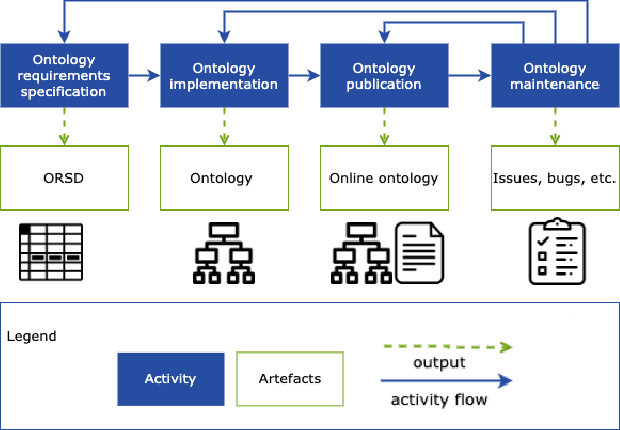
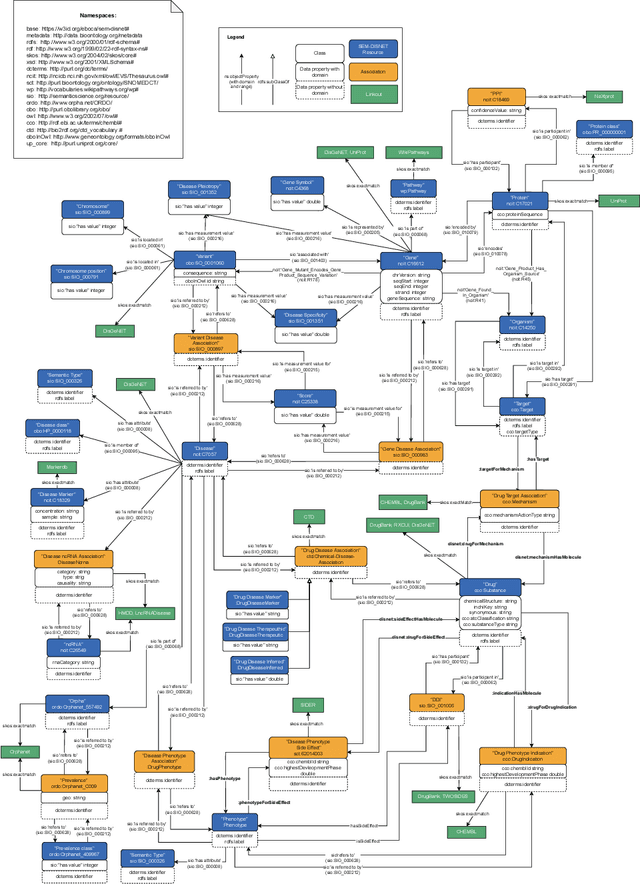
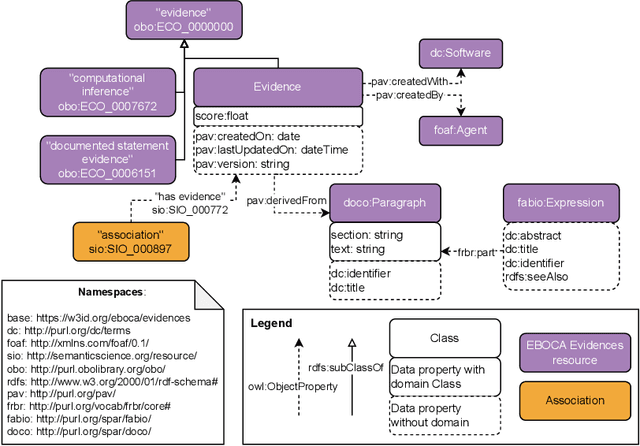
Abstract:There is a large number of online documents data sources available nowadays. The lack of structure and the differences between formats are the main difficulties to automatically extract information from them, which also has a negative impact on its use and reuse. In the biomedical domain, the DISNET platform emerged to provide researchers with a resource to obtain information in the scope of human disease networks by means of large-scale heterogeneous sources. Specifically in this domain, it is critical to offer not only the information extracted from different sources, but also the evidence that supports it. This paper proposes EBOCA, an ontology that describes (i) biomedical domain concepts and associations between them, and (ii) evidences supporting these associations; with the objective of providing an schema to improve the publication and description of evidences and biomedical associations in this domain. The ontology has been successfully evaluated to ensure there are no errors, modelling pitfalls and that it meets the previously defined functional requirements. Test data coming from a subset of DISNET and automatic association extractions from texts has been transformed according to the proposed ontology to create a Knowledge Graph that can be used in real scenarios, and which has also been used for the evaluation of the presented ontology.
Efficient Clustering from Distributions over Topics
Dec 15, 2020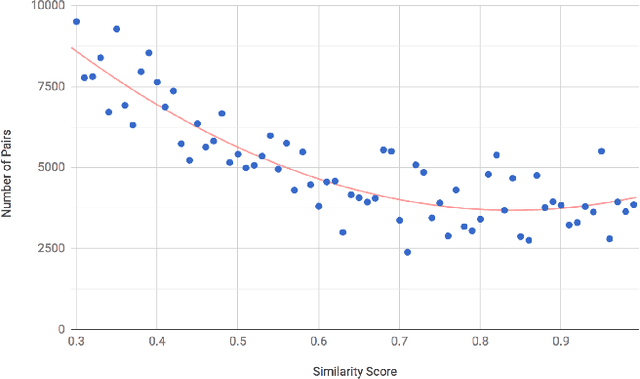
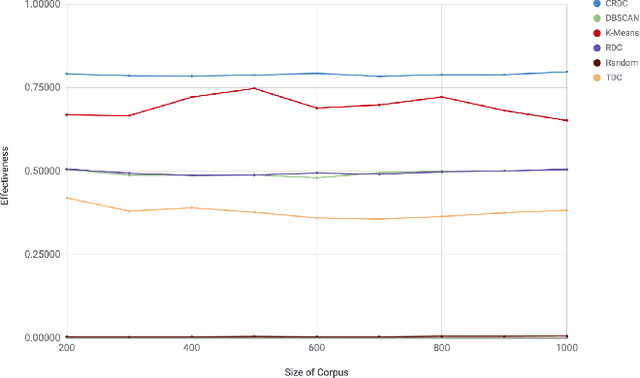
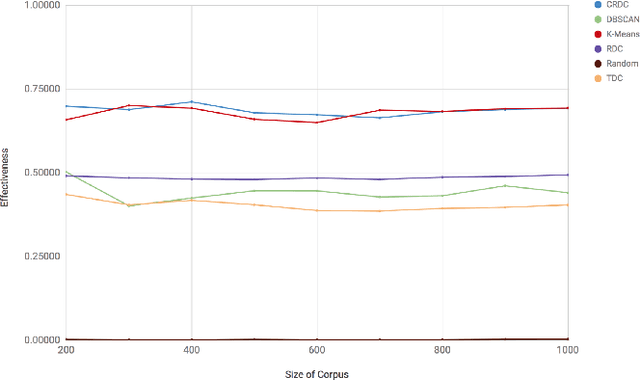
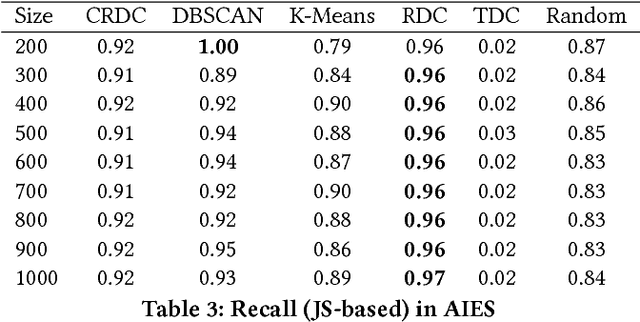
Abstract:There are many scenarios where we may want to find pairs of textually similar documents in a large corpus (e.g. a researcher doing literature review, or an R&D project manager analyzing project proposals). To programmatically discover those connections can help experts to achieve those goals, but brute-force pairwise comparisons are not computationally adequate when the size of the document corpus is too large. Some algorithms in the literature divide the search space into regions containing potentially similar documents, which are later processed separately from the rest in order to reduce the number of pairs compared. However, this kind of unsupervised methods still incur in high temporal costs. In this paper, we present an approach that relies on the results of a topic modeling algorithm over the documents in a collection, as a means to identify smaller subsets of documents where the similarity function can then be computed. This approach has proved to obtain promising results when identifying similar documents in the domain of scientific publications. We have compared our approach against state of the art clustering techniques and with different configurations for the topic modeling algorithm. Results suggest that our approach outperforms (> 0.5) the other analyzed techniques in terms of efficiency.
* Accepted at the 9th International Conference on Knowledge Capture (K-CAP 2017)
Scalable Cross-lingual Document Similarity through Language-specific Concept Hierarchies
Dec 15, 2020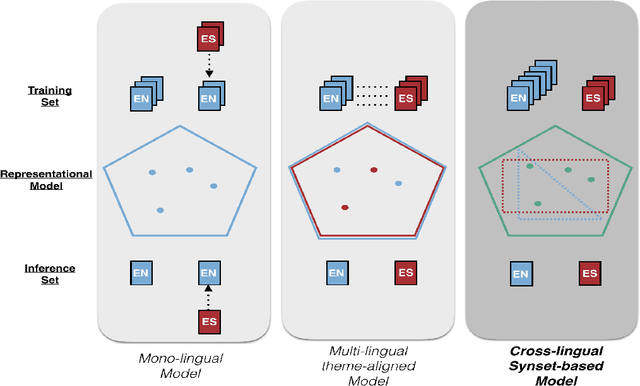
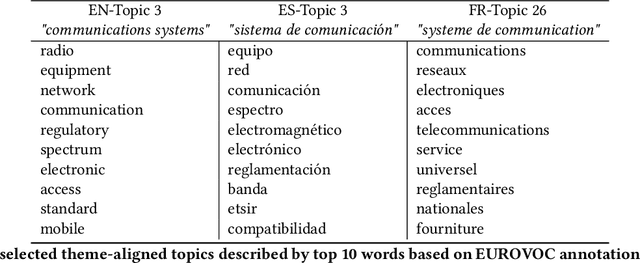
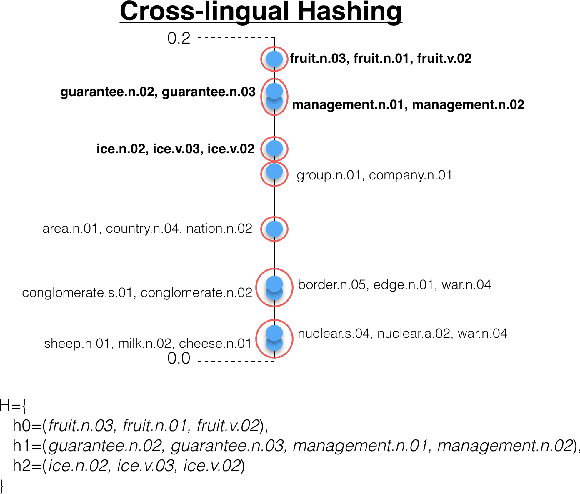
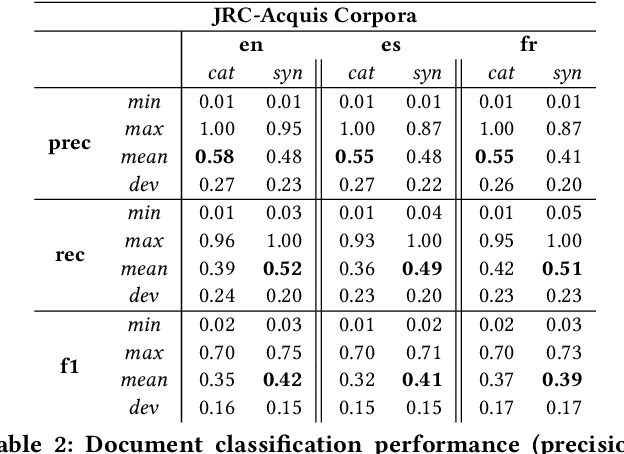
Abstract:With the ongoing growth in number of digital articles in a wider set of languages and the expanding use of different languages, we need annotation methods that enable browsing multi-lingual corpora. Multilingual probabilistic topic models have recently emerged as a group of semi-supervised machine learning models that can be used to perform thematic explorations on collections of texts in multiple languages. However, these approaches require theme-aligned training data to create a language-independent space. This constraint limits the amount of scenarios that this technique can offer solutions to train and makes it difficult to scale up to situations where a huge collection of multi-lingual documents are required during the training phase. This paper presents an unsupervised document similarity algorithm that does not require parallel or comparable corpora, or any other type of translation resource. The algorithm annotates topics automatically created from documents in a single language with cross-lingual labels and describes documents by hierarchies of multi-lingual concepts from independently-trained models. Experiments performed on the English, Spanish and French editions of JCR-Acquis corpora reveal promising results on classifying and sorting documents by similar content.
* Accepted at the 10th International Conference on Knowledge Capture (K-CAP 2019)
Drugs4Covid: Drug-driven Knowledge Exploitation based on Scientific Publications
Dec 03, 2020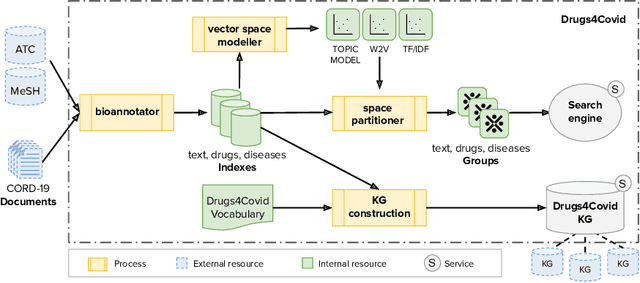
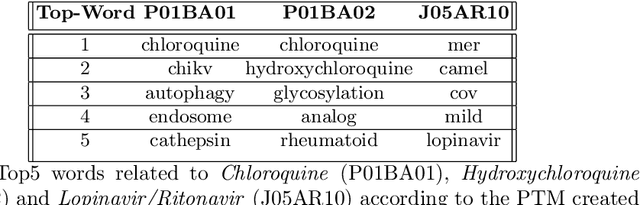

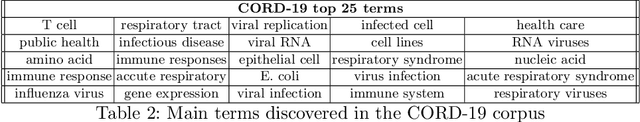
Abstract:In the absence of sufficient medication for COVID patients due to the increased demand, disused drugs have been employed or the doses of those available were modified by hospital pharmacists. Some evidences for the use of alternative drugs can be found in the existing scientific literature that could assist in such decisions. However, exploiting large corpus of documents in an efficient manner is not easy, since drugs may not appear explicitly related in the texts and could be mentioned under different brand names. Drugs4Covid combines word embedding techniques and semantic web technologies to enable a drug-oriented exploration of large medical literature. Drugs and diseases are identified according to the ATC classification and MeSH categories respectively. More than 60K articles and 2M paragraphs have been processed from the CORD-19 corpus with information of COVID-19, SARS, and other related coronaviruses. An open catalogue of drugs has been created and results are publicly available through a drug browser, a keyword-guided text explorer, and a knowledge graph.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge