Joan Llop-Palao
Biomedical and Clinical Language Models for Spanish: On the Benefits of Domain-Specific Pretraining in a Mid-Resource Scenario
Sep 17, 2021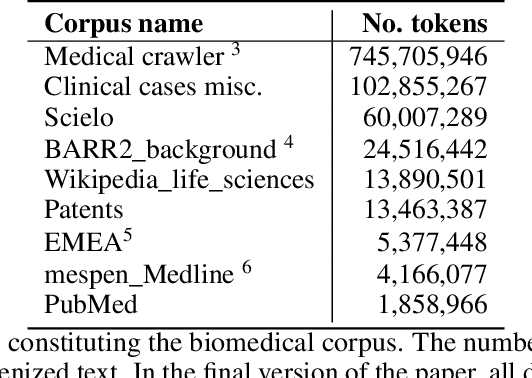
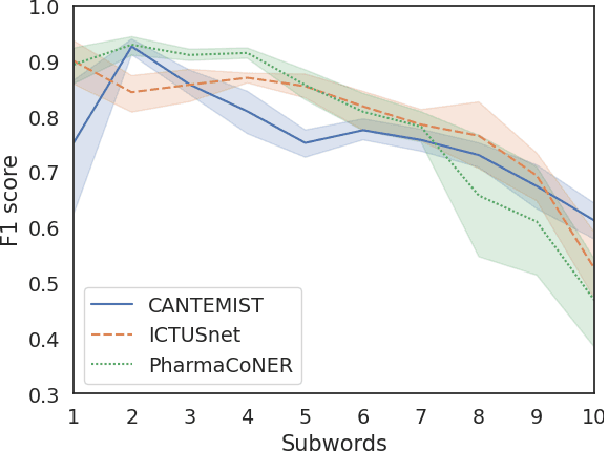
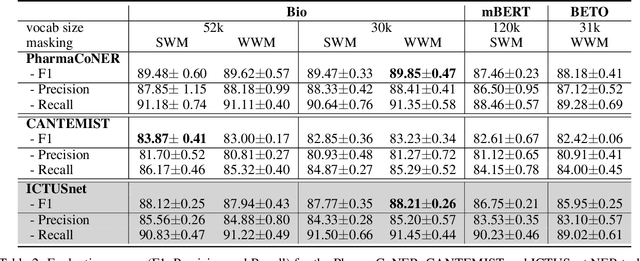
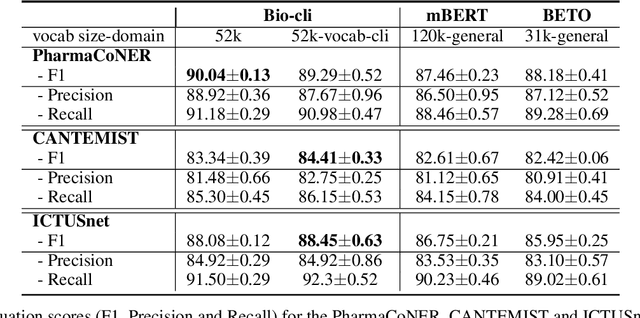
Abstract:This work presents biomedical and clinical language models for Spanish by experimenting with different pretraining choices, such as masking at word and subword level, varying the vocabulary size and testing with domain data, looking for better language representations. Interestingly, in the absence of enough clinical data to train a model from scratch, we applied mixed-domain pretraining and cross-domain transfer approaches to generate a performant bio-clinical model suitable for real-world clinical data. We evaluated our models on Named Entity Recognition (NER) tasks for biomedical documents and challenging hospital discharge reports. When compared against the competitive mBERT and BETO models, we outperform them in all NER tasks by a significant margin. Finally, we studied the impact of the model's vocabulary on the NER performances by offering an interesting vocabulary-centric analysis. The results confirm that domain-specific pretraining is fundamental to achieving higher performances in downstream NER tasks, even within a mid-resource scenario. To the best of our knowledge, we provide the first biomedical and clinical transformer-based pretrained language models for Spanish, intending to boost native Spanish NLP applications in biomedicine. Our best models are freely available in the HuggingFace hub: https://huggingface.co/BSC-TeMU.
Spanish Language Models
Aug 13, 2021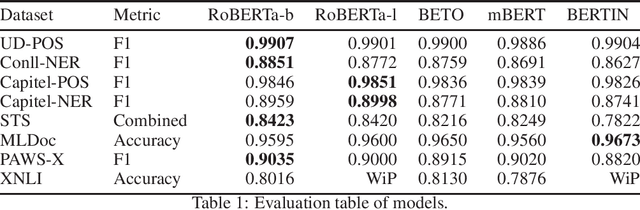
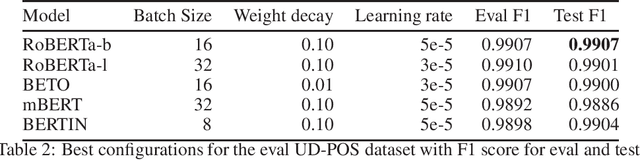
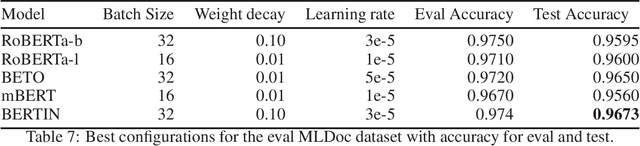
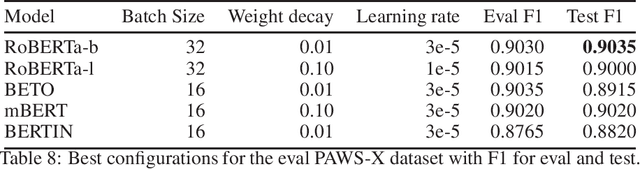
Abstract:This paper presents the Spanish RoBERTa-base and RoBERTa-large models, as well as the corresponding performance evaluations. Both models were pre-trained using the largest Spanish corpus known to date, with a total of 570GB of clean and deduplicated text processed for this work, compiled from the web crawlings performed by the National Library of Spain from 2009 to 2019. We extended the current evaluation datasets with an extractive Question Answering dataset and our models outperform the existing Spanish models across tasks and settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge