Jie-Zhi Cheng
Domain Generalization for Mammographic Image Analysis via Contrastive Learning
Apr 20, 2023Abstract:Mammographic image analysis is a fundamental problem in the computer-aided diagnosis scheme, which has recently made remarkable progress with the advance of deep learning. However, the construction of a deep learning model requires training data that are large and sufficiently diverse in terms of image style and quality. In particular, the diversity of image style may be majorly attributed to the vendor factor. However, mammogram collection from vendors as many as possible is very expensive and sometimes impractical for laboratory-scale studies. Accordingly, to further augment the generalization capability of deep learning models to various vendors with limited resources, a new contrastive learning scheme is developed. Specifically, the backbone network is firstly trained with a multi-style and multi-view unsupervised self-learning scheme for the embedding of invariant features to various vendor styles. Afterward, the backbone network is then recalibrated to the downstream tasks of mass detection, multi-view mass matching, BI-RADS classification and breast density classification with specific supervised learning. The proposed method is evaluated with mammograms from four vendors and two unseen public datasets. The experimental results suggest that our approach can effectively improve analysis performance on both seen and unseen domains, and outperforms many state-of-the-art (SOTA) generalization methods.
Image Synthesis with Disentangled Attributes for Chest X-Ray Nodule Augmentation and Detection
Jul 19, 2022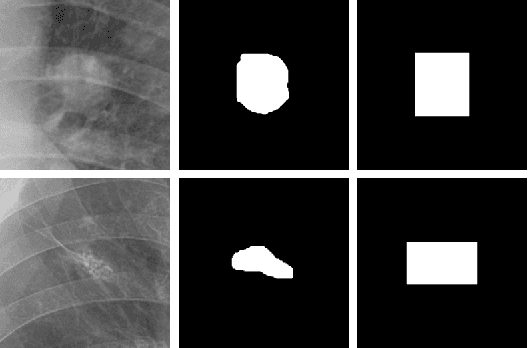
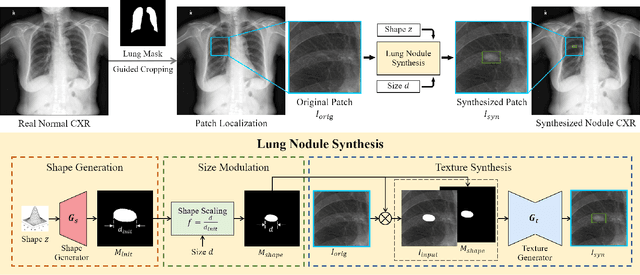
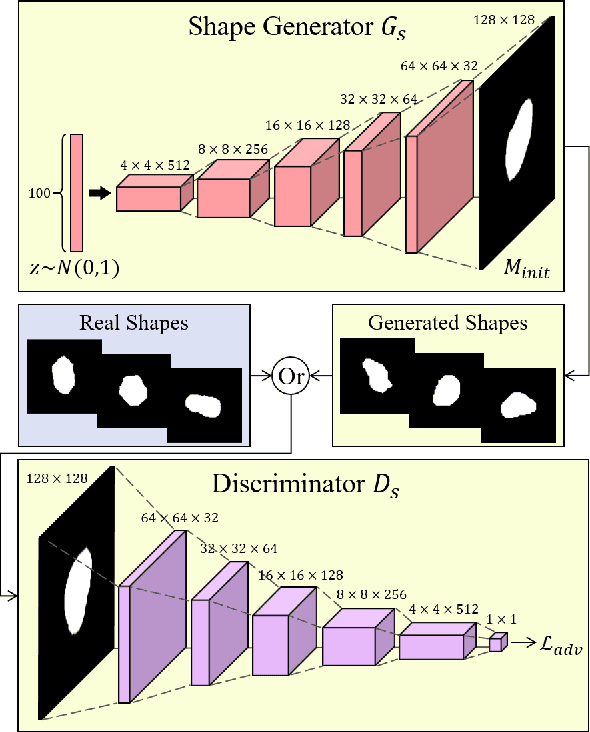
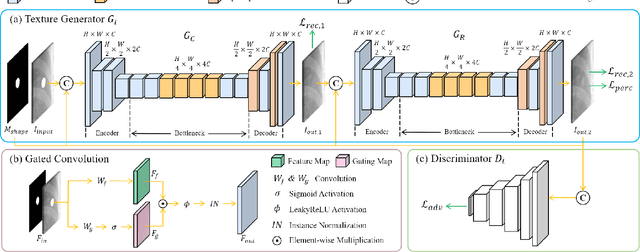
Abstract:Lung nodule detection in chest X-ray (CXR) images is common to early screening of lung cancers. Deep-learning-based Computer-Assisted Diagnosis (CAD) systems can support radiologists for nodule screening in CXR. However, it requires large-scale and diverse medical data with high-quality annotations to train such robust and accurate CADs. To alleviate the limited availability of such datasets, lung nodule synthesis methods are proposed for the sake of data augmentation. Nevertheless, previous methods lack the ability to generate nodules that are realistic with the size attribute desired by the detector. To address this issue, we introduce a novel lung nodule synthesis framework in this paper, which decomposes nodule attributes into three main aspects including shape, size, and texture, respectively. A GAN-based Shape Generator firstly models nodule shapes by generating diverse shape masks. The following Size Modulation then enables quantitative control on the diameters of the generated nodule shapes in pixel-level granularity. A coarse-to-fine gated convolutional Texture Generator finally synthesizes visually plausible nodule textures conditioned on the modulated shape masks. Moreover, we propose to synthesize nodule CXR images by controlling the disentangled nodule attributes for data augmentation, in order to better compensate for the nodules that are easily missed in the detection task. Our experiments demonstrate the enhanced image quality, diversity, and controllability of the proposed lung nodule synthesis framework. We also validate the effectiveness of our data augmentation on greatly improving nodule detection performance.
Learning Hierarchical Attention for Weakly-supervised Chest X-Ray Abnormality Localization and Diagnosis
Dec 23, 2021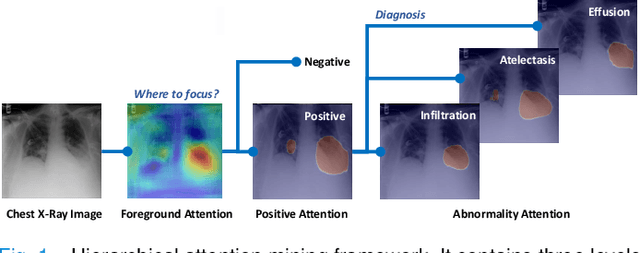
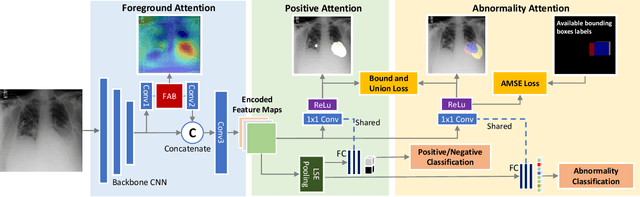
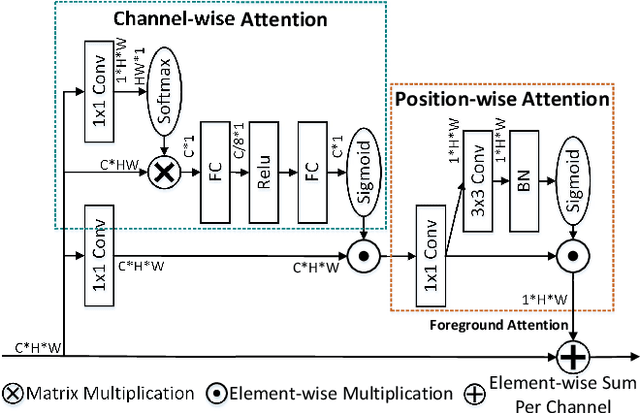
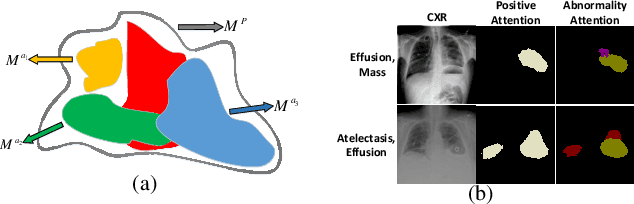
Abstract:We consider the problem of abnormality localization for clinical applications. While deep learning has driven much recent progress in medical imaging, many clinical challenges are not fully addressed, limiting its broader usage. While recent methods report high diagnostic accuracies, physicians have concerns trusting these algorithm results for diagnostic decision-making purposes because of a general lack of algorithm decision reasoning and interpretability. One potential way to address this problem is to further train these models to localize abnormalities in addition to just classifying them. However, doing this accurately will require a large amount of disease localization annotations by clinical experts, a task that is prohibitively expensive to accomplish for most applications. In this work, we take a step towards addressing these issues by means of a new attention-driven weakly supervised algorithm comprising a hierarchical attention mining framework that unifies activation- and gradient-based visual attention in a holistic manner. Our key algorithmic innovations include the design of explicit ordinal attention constraints, enabling principled model training in a weakly-supervised fashion, while also facilitating the generation of visual-attention-driven model explanations by means of localization cues. On two large-scale chest X-ray datasets (NIH ChestX-ray14 and CheXpert), we demonstrate significant localization performance improvements over the current state of the art while also achieving competitive classification performance. Our code is available on https://github.com/oyxhust/HAM.
Domain Generalization for Mammography Detection via Multi-style and Multi-view Contrastive Learning
Nov 21, 2021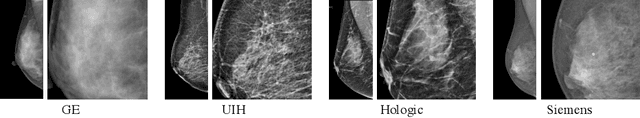
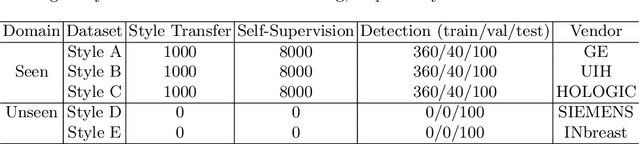
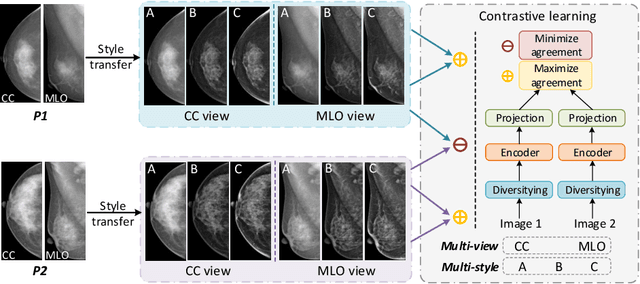
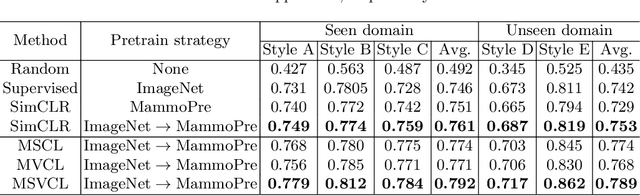
Abstract:Lesion detection is a fundamental problem in the computer-aided diagnosis scheme for mammography. The advance of deep learning techniques have made a remarkable progress for this task, provided that the training data are large and sufficiently diverse in terms of image style and quality. In particular, the diversity of image style may be majorly attributed to the vendor factor. However, the collection of mammograms from vendors as many as possible is very expensive and sometimes impractical for laboratory-scale studies. Accordingly, to further augment the generalization capability of deep learning model to various vendors with limited resources, a new contrastive learning scheme is developed. Specifically, the backbone network is firstly trained with a multi-style and multi-view unsupervised self-learning scheme for the embedding of invariant features to various vendor-styles. Afterward, the backbone network is then recalibrated to the downstream task of lesion detection with the specific supervised learning. The proposed method is evaluated with mammograms from four vendors and one unseen public dataset. The experimental results suggest that our approach can effectively improve detection performance on both seen and unseen domains, and outperforms many state-of-the-art (SOTA) generalization methods.
* Pages 98-108
mr2NST: Multi-Resolution and Multi-Reference Neural Style Transfer for Mammography
May 25, 2020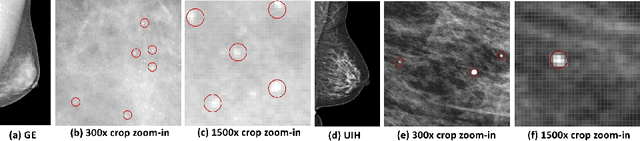

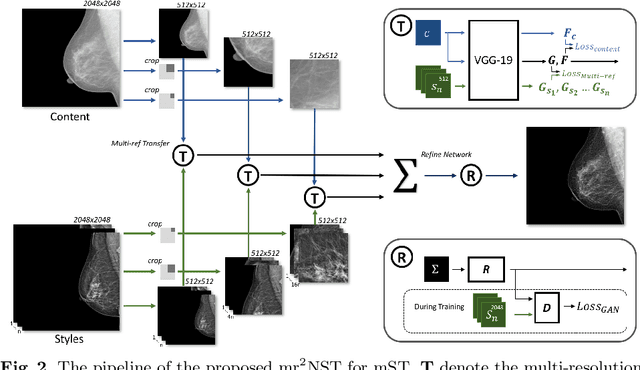
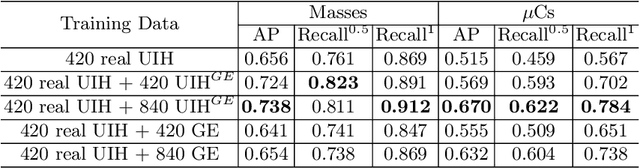
Abstract:Computer-aided diagnosis with deep learning techniques has been shown to be helpful for the diagnosis of the mammography in many clinical studies. However, the image styles of different vendors are very distinctive, and there may exist domain gap among different vendors that could potentially compromise the universal applicability of one deep learning model. In this study, we explicitly address style variety issue with the proposed multi-resolution and multi-reference neural style transfer (mr2NST) network. The mr2NST can normalize the styles from different vendors to the same style baseline with very high resolution. We illustrate that the image quality of the transferred images is comparable to the quality of original images of the target domain (vendor) in terms of NIMA scores. Meanwhile, the mr2NST results are also shown to be helpful for the lesion detection in mammograms.
Automatic 3D Cardiovascular MR Segmentation with Densely-Connected Volumetric ConvNets
Aug 02, 2017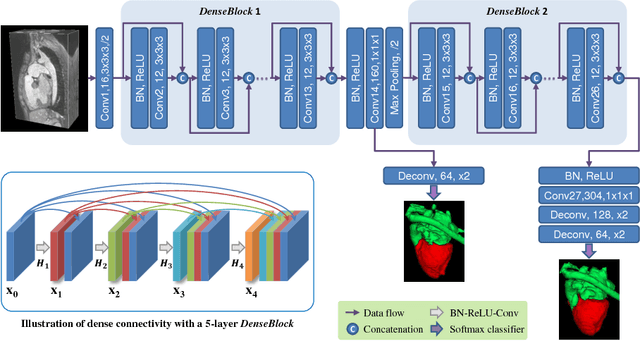
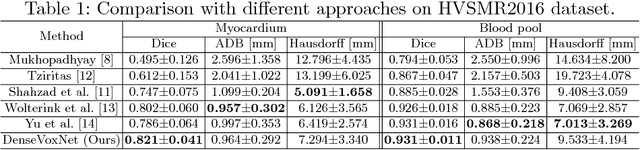
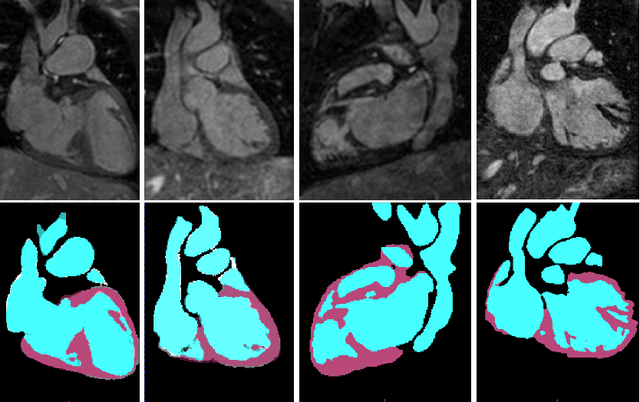
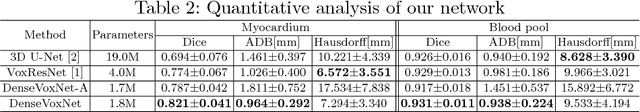
Abstract:Automatic and accurate whole-heart and great vessel segmentation from 3D cardiac magnetic resonance (MR) images plays an important role in the computer-assisted diagnosis and treatment of cardiovascular disease. However, this task is very challenging due to ambiguous cardiac borders and large anatomical variations among different subjects. In this paper, we propose a novel densely-connected volumetric convolutional neural network, referred as DenseVoxNet, to automatically segment the cardiac and vascular structures from 3D cardiac MR images. The DenseVoxNet adopts the 3D fully convolutional architecture for effective volume-to-volume prediction. From the learning perspective, our DenseVoxNet has three compelling advantages. First, it preserves the maximum information flow between layers by a densely-connected mechanism and hence eases the network training. Second, it avoids learning redundant feature maps by encouraging feature reuse and hence requires fewer parameters to achieve high performance, which is essential for medical applications with limited training data. Third, we add auxiliary side paths to strengthen the gradient propagation and stabilize the learning process. We demonstrate the effectiveness of DenseVoxNet by comparing it with the state-of-the-art approaches from HVSMR 2016 challenge in conjunction with MICCAI, and our network achieves the best dice coefficient. We also show that our network can achieve better performance than other 3D ConvNets but with fewer parameters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge