Houman Owhadi
Solving Functional PDEs with Gaussian Processes and Applications to Functional Renormalization Group Equations
Dec 24, 2025Abstract:We present an operator learning framework for solving non-perturbative functional renormalization group equations, which are integro-differential equations defined on functionals. Our proposed approach uses Gaussian process operator learning to construct a flexible functional representation formulated directly on function space, making it independent of a particular equation or discretization. Our method is flexible, and can apply to a broad range of functional differential equations while still allowing for the incorporation of physical priors in either the prior mean or the kernel design. We demonstrate the performance of our method on several relevant equations, such as the Wetterich and Wilson--Polchinski equations, showing that it achieves equal or better performance than existing approximations such as the local-potential approximation, while being significantly more flexible. In particular, our method can handle non-constant fields, making it promising for the study of more complex field configurations, such as instantons.
Data-Efficient Kernel Methods for Learning Differential Equations and Their Solution Operators: Algorithms and Error Analysis
Mar 02, 2025Abstract:We introduce a novel kernel-based framework for learning differential equations and their solution maps that is efficient in data requirements, in terms of solution examples and amount of measurements from each example, and computational cost, in terms of training procedures. Our approach is mathematically interpretable and backed by rigorous theoretical guarantees in the form of quantitative worst-case error bounds for the learned equation. Numerical benchmarks demonstrate significant improvements in computational complexity and robustness while achieving one to two orders of magnitude improvements in terms of accuracy compared to state-of-the-art algorithms.
Solving Roughly Forced Nonlinear PDEs via Misspecified Kernel Methods and Neural Networks
Jan 29, 2025Abstract:We consider the use of Gaussian Processes (GPs) or Neural Networks (NNs) to numerically approximate the solutions to nonlinear partial differential equations (PDEs) with rough forcing or source terms, which commonly arise as pathwise solutions to stochastic PDEs. Kernel methods have recently been generalized to solve nonlinear PDEs by approximating their solutions as the maximum a posteriori estimator of GPs that are conditioned to satisfy the PDE at a finite set of collocation points. The convergence and error guarantees of these methods, however, rely on the PDE being defined in a classical sense and its solution possessing sufficient regularity to belong to the associated reproducing kernel Hilbert space. We propose a generalization of these methods to handle roughly forced nonlinear PDEs while preserving convergence guarantees with an oversmoothing GP kernel that is misspecified relative to the true solution's regularity. This is achieved by conditioning a regular GP to satisfy the PDE with a modified source term in a weak sense (when integrated against a finite number of test functions). This is equivalent to replacing the empirical $L^2$-loss on the PDE constraint by an empirical negative-Sobolev norm. We further show that this loss function can be used to extend physics-informed neural networks (PINNs) to stochastic equations, thereby resulting in a new NN-based variant termed Negative Sobolev Norm-PINN (NeS-PINN).
Kernel Methods for the Approximation of the Eigenfunctions of the Koopman Operator
Dec 21, 2024



Abstract:The Koopman operator provides a linear framework to study nonlinear dynamical systems. Its spectra offer valuable insights into system dynamics, but the operator can exhibit both discrete and continuous spectra, complicating direct computations. In this paper, we introduce a kernel-based method to construct the principal eigenfunctions of the Koopman operator without explicitly computing the operator itself. These principal eigenfunctions are associated with the equilibrium dynamics, and their eigenvalues match those of the linearization of the nonlinear system at the equilibrium point. We exploit the structure of the principal eigenfunctions by decomposing them into linear and nonlinear components. The linear part corresponds to the left eigenvector of the system's linearization at the equilibrium, while the nonlinear part is obtained by solving a partial differential equation (PDE) using kernel methods. Our approach avoids common issues such as spectral pollution and spurious eigenvalues, which can arise in previous methods. We demonstrate the effectiveness of our algorithm through numerical examples.
Toward Efficient Kernel-Based Solvers for Nonlinear PDEs
Oct 15, 2024Abstract:This paper introduces a novel kernel learning framework toward efficiently solving nonlinear partial differential equations (PDEs). In contrast to the state-of-the-art kernel solver that embeds differential operators within kernels, posing challenges with a large number of collocation points, our approach eliminates these operators from the kernel. We model the solution using a standard kernel interpolation form and differentiate the interpolant to compute the derivatives. Our framework obviates the need for complex Gram matrix construction between solutions and their derivatives, allowing for a straightforward implementation and scalable computation. As an instance, we allocate the collocation points on a grid and adopt a product kernel, which yields a Kronecker product structure in the interpolation. This structure enables us to avoid computing the full Gram matrix, reducing costs and scaling efficiently to a large number of collocation points. We provide a proof of the convergence and rate analysis of our method under appropriate regularity assumptions. In numerical experiments, we demonstrate the advantages of our method in solving several benchmark PDEs.
Model aggregation: minimizing empirical variance outperforms minimizing empirical error
Sep 25, 2024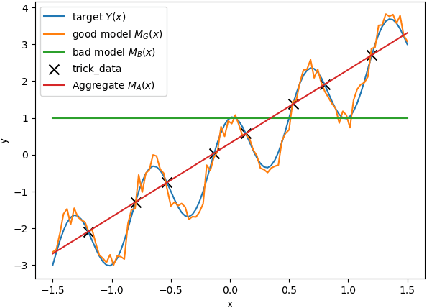
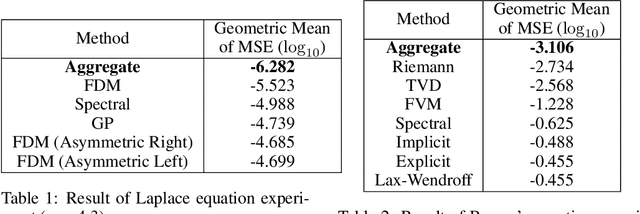
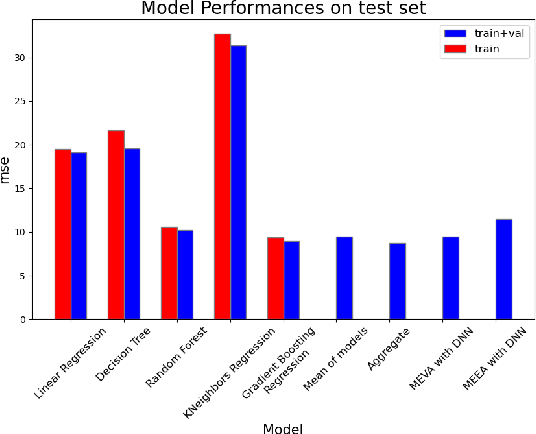
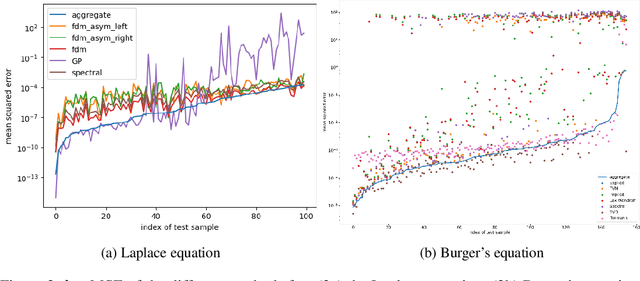
Abstract:Whether deterministic or stochastic, models can be viewed as functions designed to approximate a specific quantity of interest. We propose a data-driven framework that aggregates predictions from diverse models into a single, more accurate output. This aggregation approach exploits each model's strengths to enhance overall accuracy. It is non-intrusive - treating models as black-box functions - model-agnostic, requires minimal assumptions, and can combine outputs from a wide range of models, including those from machine learning and numerical solvers. We argue that the aggregation process should be point-wise linear and propose two methods to find an optimal aggregate: Minimal Error Aggregation (MEA), which minimizes the aggregate's prediction error, and Minimal Variance Aggregation (MVA), which minimizes its variance. While MEA is inherently more accurate when correlations between models and the target quantity are perfectly known, Minimal Empirical Variance Aggregation (MEVA), an empirical version of MVA - consistently outperforms Minimal Empirical Error Aggregation (MEEA), the empirical counterpart of MEA, when these correlations must be estimated from data. The key difference is that MEVA constructs an aggregate by estimating model errors, while MEEA treats the models as features for direct interpolation of the quantity of interest. This makes MEEA more susceptible to overfitting and poor generalization, where the aggregate may underperform individual models during testing. We demonstrate the versatility and effectiveness of our framework in various applications, such as data science and partial differential equations, showing how it successfully integrates traditional solvers with machine learning models to improve both robustness and accuracy.
Operator Learning with Gaussian Processes
Sep 06, 2024


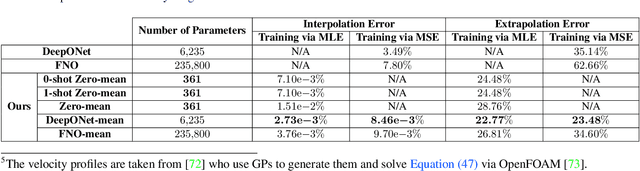
Abstract:Operator learning focuses on approximating mappings $\mathcal{G}^\dagger:\mathcal{U} \rightarrow\mathcal{V}$ between infinite-dimensional spaces of functions, such as $u: \Omega_u\rightarrow\mathbb{R}$ and $v: \Omega_v\rightarrow\mathbb{R}$. This makes it particularly suitable for solving parametric nonlinear partial differential equations (PDEs). While most machine learning methods for operator learning rely on variants of deep neural networks (NNs), recent studies have shown that Gaussian Processes (GPs) are also competitive while offering interpretability and theoretical guarantees. In this paper, we introduce a hybrid GP/NN-based framework for operator learning that leverages the strengths of both methods. Instead of approximating the function-valued operator $\mathcal{G}^\dagger$, we use a GP to approximate its associated real-valued bilinear form $\widetilde{\mathcal{G}}^\dagger: \mathcal{U}\times\mathcal{V}^*\rightarrow\mathbb{R}.$ This bilinear form is defined by $\widetilde{\mathcal{G}}^\dagger(u,\varphi) := [\varphi,\mathcal{G}^\dagger(u)],$ which allows us to recover the operator $\mathcal{G}^\dagger$ through $\mathcal{G}^\dagger(u)(y)=\widetilde{\mathcal{G}}^\dagger(u,\delta_y).$ The GP mean function can be zero or parameterized by a neural operator and for each setting we develop a robust training mechanism based on maximum likelihood estimation (MLE) that can optionally leverage the physics involved. Numerical benchmarks show that (1) it improves the performance of a base neural operator by using it as the mean function of a GP, and (2) it enables zero-shot data-driven models for accurate predictions without prior training. Our framework also handles multi-output operators where $\mathcal{G}^\dagger:\mathcal{U} \rightarrow\prod_{s=1}^S\mathcal{V}^s$, and benefits from computational speed-ups via product kernel structures and Kronecker product matrix representations.
Kernel Sum of Squares for Data Adapted Kernel Learning of Dynamical Systems from Data: A global optimization approach
Aug 12, 2024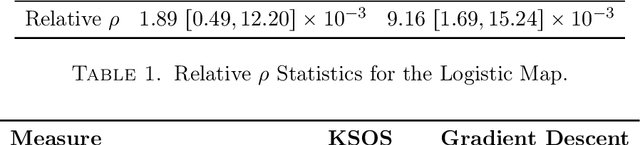
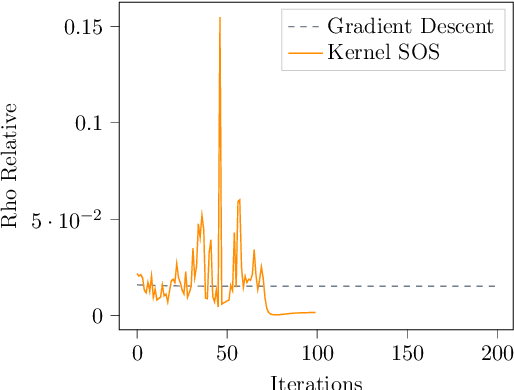
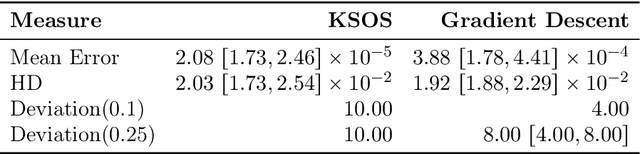
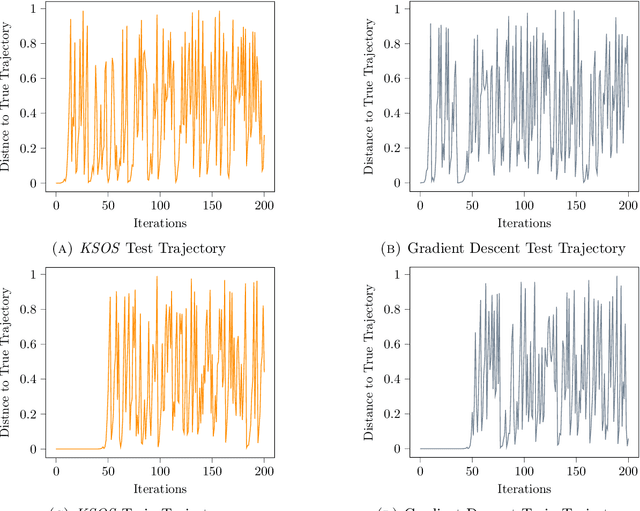
Abstract:This paper examines the application of the Kernel Sum of Squares (KSOS) method for enhancing kernel learning from data, particularly in the context of dynamical systems. Traditional kernel-based methods, despite their theoretical soundness and numerical efficiency, frequently struggle with selecting optimal base kernels and parameter tuning, especially with gradient-based methods prone to local optima. KSOS mitigates these issues by leveraging a global optimization framework with kernel-based surrogate functions, thereby achieving more reliable and precise learning of dynamical systems. Through comprehensive numerical experiments on the Logistic Map, Henon Map, and Lorentz System, KSOS is shown to consistently outperform gradient descent in minimizing the relative-$\rho$ metric and improving kernel accuracy. These results highlight KSOS's effectiveness in predicting the behavior of chaotic dynamical systems, demonstrating its capability to adapt kernels to underlying dynamics and enhance the robustness and predictive power of kernel-based approaches, making it a valuable asset for time series analysis in various scientific fields.
Gaussian Measures Conditioned on Nonlinear Observations: Consistency, MAP Estimators, and Simulation
May 21, 2024



Abstract:The article presents a systematic study of the problem of conditioning a Gaussian random variable $\xi$ on nonlinear observations of the form $F \circ \phi(\xi)$ where $\phi: \mathcal{X} \to \mathbb{R}^N$ is a bounded linear operator and $F$ is nonlinear. Such problems arise in the context of Bayesian inference and recent machine learning-inspired PDE solvers. We give a representer theorem for the conditioned random variable $\xi \mid F\circ \phi(\xi)$, stating that it decomposes as the sum of an infinite-dimensional Gaussian (which is identified analytically) as well as a finite-dimensional non-Gaussian measure. We also introduce a novel notion of the mode of a conditional measure by taking the limit of the natural relaxation of the problem, to which we can apply the existing notion of maximum a posteriori estimators of posterior measures. Finally, we introduce a variant of the Laplace approximation for the efficient simulation of the aforementioned conditioned Gaussian random variables towards uncertainty quantification.
Kolmogorov n-Widths for Multitask Physics-Informed Machine Learning (PIML) Methods: Towards Robust Metrics
Feb 16, 2024



Abstract:Physics-informed machine learning (PIML) as a means of solving partial differential equations (PDE) has garnered much attention in the Computational Science and Engineering (CS&E) world. This topic encompasses a broad array of methods and models aimed at solving a single or a collection of PDE problems, called multitask learning. PIML is characterized by the incorporation of physical laws into the training process of machine learning models in lieu of large data when solving PDE problems. Despite the overall success of this collection of methods, it remains incredibly difficult to analyze, benchmark, and generally compare one approach to another. Using Kolmogorov n-widths as a measure of effectiveness of approximating functions, we judiciously apply this metric in the comparison of various multitask PIML architectures. We compute lower accuracy bounds and analyze the model's learned basis functions on various PDE problems. This is the first objective metric for comparing multitask PIML architectures and helps remove uncertainty in model validation from selective sampling and overfitting. We also identify avenues of improvement for model architectures, such as the choice of activation function, which can drastically affect model generalization to "worst-case" scenarios, which is not observed when reporting task-specific errors. We also incorporate this metric into the optimization process through regularization, which improves the models' generalizability over the multitask PDE problem.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge